SWOT Software Design Document (SDD)
Introduction
Purpose
This document provides the high level design information to understand the architecture and design of the core HySDS framework. It is not intended to replace reading the code for details.
Background
The primary responsibility of a science data system (SDS) is to ingest science data as input and process them into higher level science data products as output.
The SDS black box obviously provides more capability than what's depicted in the figure above and so the point is that any and all capability evolves from this fundamental principle. In 1978 one of the earliest Earth-observing satellites, NASA’s SEASAT, was launched which gave rise to one of the earliest SDS implementations at NASA’s Jet Propulsion Laboratory (JPL), the Interim Digital SAR Processor (IDP).
The development of the IDP was driven by the throughput requirement of “approximately one SEASAT SAR frame (100 km x 100 km coverage, 25 m resolution, 4 looks) per ten hours of processing time” (Wu et al. 1981). The overall average throughput rate of the IDP SDS was approximately 3.5 SEASAT SAR frames per week and the infrastructure utilized by the IDP to fulfill those requirements was maintainable on-premise.
Over the years technological advancements in observation platforms, resource availability, instrument sensitivities, communications downlink bandwidth, and improved science processing algorithms have resulted in increasingly larger science data volumes and data rates. There was also an increased demand in high-throughput, lower-latency, time-critical data production. Subsequent efforts to improve the performance of the on-premise SDS include parallelization of compute algorithms (Leung et al. 1997), scaling compute horizontally to HPC/HEC resources such as Pleiades (LaHaye 2012), and leveraging GPUs for high-performance arithmetic operations (Cohen and Agram 2017). Despite these efforts, the ever increasing requirements of science data volume, latency and throughput translates to a major problem for the on-premise SDS: scaling on-premise resources and their associated costs.
Before the advent of the cloud paradigm, the machinery for the SDS had mostly been hosted and operated on-premise at data centers owned and operated by the respective stakeholders. In this era of big data, larger requirements and cost constraints are forcing functions for moving the SDS as well as other associated data systems from the enterprise to the cloud. HySDS, the Hybrid Cloud Science Data System, was born in 2009 to fulfill this need for the Advanced Rapid Imaging and Analysis project (ARIA) and is subsequently used in various other projects today (SMAP, SWOT, NISAR, ARIA-SG, WVCC).
Overview
HySDS is intended to support a variety of functions in support of science data processing. The functions that HySDS is intended to perform include the following:
- Automate deployment onto on-premise resources and non-specific cloud platforms
- Allow integration of hybrid, multi-cloud, and HPC/HEC compute resources under a single deployment
- Scalably ingest, catalog and store data and ancillary datasets enabling science data processing
- Interface with science data processors encapsulated in containers
- Provide workflow capabilities to chain science data processors together
- Automate scalable science data processing to produce downstream science data products
- Store and catalog science data products and their associated files
- Provide search capability and access to science data products using the catalog
- Make catalogued data products accessible and searchable
- Provide on-demand and periodic reporting capability
- Track and visualize science data processing metrics such as execution duration, number of failed jobs, products produced and so on
- Track and visualize infrastructure metrics such as CPU usage, memory usage, storage usage an so on
The diagram below depicts this end-to-end functionality.
Design Philosophy, Assumptions and Constraints
Design Philosophy
Born out of R&D projects, the HySDS design does not explicitly subscribe to any predefined set of principles. This may change in the future as the formalization of a HySDS develop/user community continues to evolve and take shape. In general though, the development of HySDS core components has usually followed the following set of principles conducive to agile development:
- KISS (Keep it simple, silly) - Use the simplest solution that solves the problem.
- Eat your own dog food - reuse solutions that are already provided by the system.
- YAGNI (You aren't going to need it) - Build it only when you need it.
- Crawl, walk, run - Do iterative, agile development.
- Automate - Everything must be automated: tests, releases, configuration management, etc.
- ROI (Return on investment) - Pay attention to what will make the most impact.
- Know your users - Develop features that you know will be used.
- Design and test features independently.
- Beware Google Envy - Don't bring features or solutions that you won't need just because it's the sexiest thing out there.
- MVP (Minimum viable product) - Design a feature that supports you use case and conforms to the KISS principle.
- Expect failure at scale - All the testing in the world will not expose issues that only happen at scale. Expect them and mitigate using techniques like exponential backoff.
Design Assumptions
The HySDS design involves the following assumptions:
- Industry standards (protocols, formats, security practices, etc.) should be utilized, e.g. HTTPS/SSL, JSON, XML, OpenAPI.
- HySDS must run on a 64-bit Linux operating system.
- HySDS must be platform and cloud vendor neutral, e.g. works on bare-metal, VMWare, OpenStack, AWS, GCP, Azure, HPC/HEC.
- HySDS must support long-term operations.
- HySDS must support performance intensive and time constrained processes.
- HySDS users may develop project specific adaptations to satisfy their specific needs.
Design Constraints
The HySDS software design decisions considered the following constraints:
- The implementation of HySDS should leverage on existing open-source software packages and modules as third party software.
- Capabilities of third party software - take advantage of existing capabilities and augment with custom development.
- Avoid forking third party software.
Design Framework
Architecture Overview
Core Concepts
The architecture of HySDS is described in this section through multiple views. However before diving into them let's define some core concepts that we will use throughout this technical specification document. In HySDS, a cluster is a logical grouping composed of the following 6 subsystems.
| subsystem | codename | description |
|---|---|---|
| Resource Management | mozart | Manages and coordinates all resources in the cluster. |
| Discovery | grq | Manages the storage and catalog of all datasets produced by the cluster. |
| Metrics/Analytics | metrics | Stores and visualizes metrics produced by various subsystems in the cluster. |
| Worker/Compute | verdi | Workhorse of the cluster that executes work to be done which may include producing datasets. |
| Helper | factotum | Specialized worker (verdi) that performs various core system jobs. |
| CI/CD | ci | Optional subsystem that enables development, integration, and deployment of job types into the cluster. |
Interesting note: many of the codenames used in HySDS were derived from its birth in the ARIA project (Advanced Rapid Imaging and Analysis). The word aria is defined as "a long accompanied song for a solo voice, typically one in an opera or oratorio" thus codenames were derived from this central theme. Bravo!
For more detail on each subsystem, see the HySDS Subsystem section below.
HySDS is built on Celery, a distributed task execution framework. Celery introduces the concepts of tasks and workers and HySDS essentially overlays a value-added framework to extend Celery's task to the HySDS concept of a job. The figure below provides an overview of these core concepts.
For an introduction to Celery, refer to the documentation at http://docs.celeryproject.org/en/latest/getting-started/introduction.html.
In short, Celery provides a way to define a python function that can be executed on a remote worker. It utilizes RabbitMQ as its message broker and Redis as the result backend. Producers submit tasks to the message broker which places them on queues and Consumers pull tasks from these queues to execute. For example, the following python code is a simple definition of a Celery task:
@app.task(acks_late=True)
def add(x, y):
return x + y
To submit this task to the message broker (assuming all pertinent Celery configuration parameters have been set, i.e. RabbitMQ broker url), a producer can just call the apply_async() method:
add.apply_async(1, 1, queue="math-queue")
Separately a consumer (the Celery worker server/daemon) can listen to that specific queue, execute tasks it pulls off of it, and save the results to Redis:
celery worker -Q math-queue
The following state diagram shows the different states that Celery tasks and workers can take.
The following sequence diagram provides the detailed interactions between the producer, consumer, message broker, and result backend:
It is left up to the producer to retrieve the status of the task and any results that were written to the result backend.
Reliability
The reliability of Celery to queue, pull, execute, and retrieve results is dependent on the message broker, RabbitMQ. RabbitMQ's resiliency equates to the avoidance of message loss. This means the broker must be able to handle restarts, hardware failure, crashes, and network failures. For more in-depth information on RabbitMQ's reliability, refer to the documentation at https://www.rabbitmq.com/reliability.html. In short, RabbitMQ utilizes a system of acknowledgments and confirms to ensure message delivery and conforms to AMQP 0-9-1 to provide a heartbeat feature to help detect disrupted connections and other network failures.
As stated before, HySDS overlays a value-added framework to extend Celery's task to the concept of a HySDS job. To put it succinctly, HySDS provides a Celery task (python function) that provides value-added functionality necessary to meet the functional requirements of the SDS (science data system):
@app.task(acks_late=True)
def run_job(job_payload):
# pre-process SDS job, e.g. download inputs files
...
# execute SDS job, e.g. process input files
...
# post-process SDS job, e.g. publish output files
...
More detail on HySDS job execution is provided in the following sections.
Architectural Scenarios
Let's step back up a level and look at the architecture of HySDS from multiple views. For HySDS, the architecture stakeholders are as follows:
- System Administrators
- Developers
- Operators
- Applications (workflow, schedulers)
- Users
The use case diagram above shows the primary operations scenario of HySDS. These main operations scenarios include the following:
- Submission, execution and monitoring of jobs
- Management (CRUD) of job triggers
- Accessing datasets produced by jobs
- View job, worker and dataset metrics/reports
HySDS software lifecycle scenarios are also depicted above in addition to the operations scenarios. These include:
- Development, maintenance and testing of HySDS core capability
- Deployment of HySDS
- Development of project-specific adaptations
Technology Stack
The following diagram identifies the major technologies and software components utilized by HySDS. See the implementation diagram in a later section to view the dependency tree of HySDS core components to 3rd party software.
Behavioral View
As stated previously, HySDS overlays a value-added framework to extend Celery's task to the concept of a HySDS job. This behavioral view of the architecture describes the end-to-end interactions between subsystem components during job execution. The sequence diagram depicts the recursive nature of HySDS job execution and the high-level interactions taking place at job submission through completion and trigger rule evaluation.
The following state diagram shows the different states that HySDS jobs can take:
The following state diagram shows the coupled task and job state transitions that occur over the lifecycle of a job.
More detail on HySDS job execution is provided in subsequent sections.
Structural View
The structural view of the architecture identifies the major subsystems of HySDS and their relation to the 5 core functions of execution, resource management, data management, metrics, and CI/CD. In particular the diagram depicts the mapping of each HySDS core component to these functions and where any functions are shared amongst the subsystems.
Implementation View
This view of the HySDS architecture shows the organization and dependencies of the python package code base. HySDS core packages are divided into 7 logical groupings: execution core, resource management core, data management core, deployment core, metrics core, helper core, and CI/CD core. These groupings consist of a collection of github repositories (python packages, flask applications, scripts, etc.) that together contribute to the core functionality of the grouping. The figure below also shows HySDS software dependencies to 3rd party software.
Environment View
This environment/deployment view depicts the topology of a HySDS cluster as deployed on a specific cloud-vendor, AWS. In general, the same topology will be utilized should the cluster be deployed on Google Cloud Platform, Microsoft Azure, OpenStack, Kubernetes, or on-premise bare-metal machines.
HySDS Subsystems
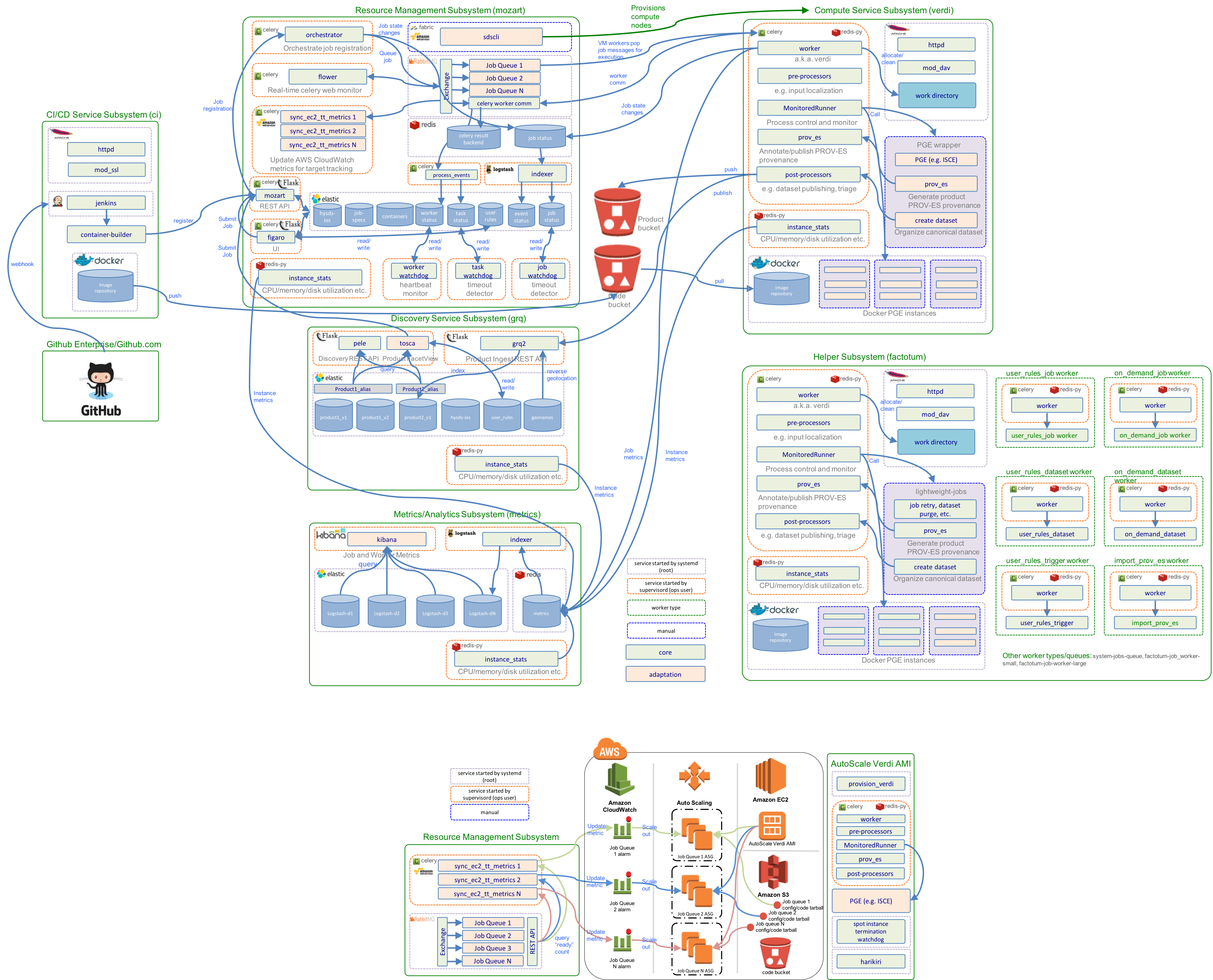
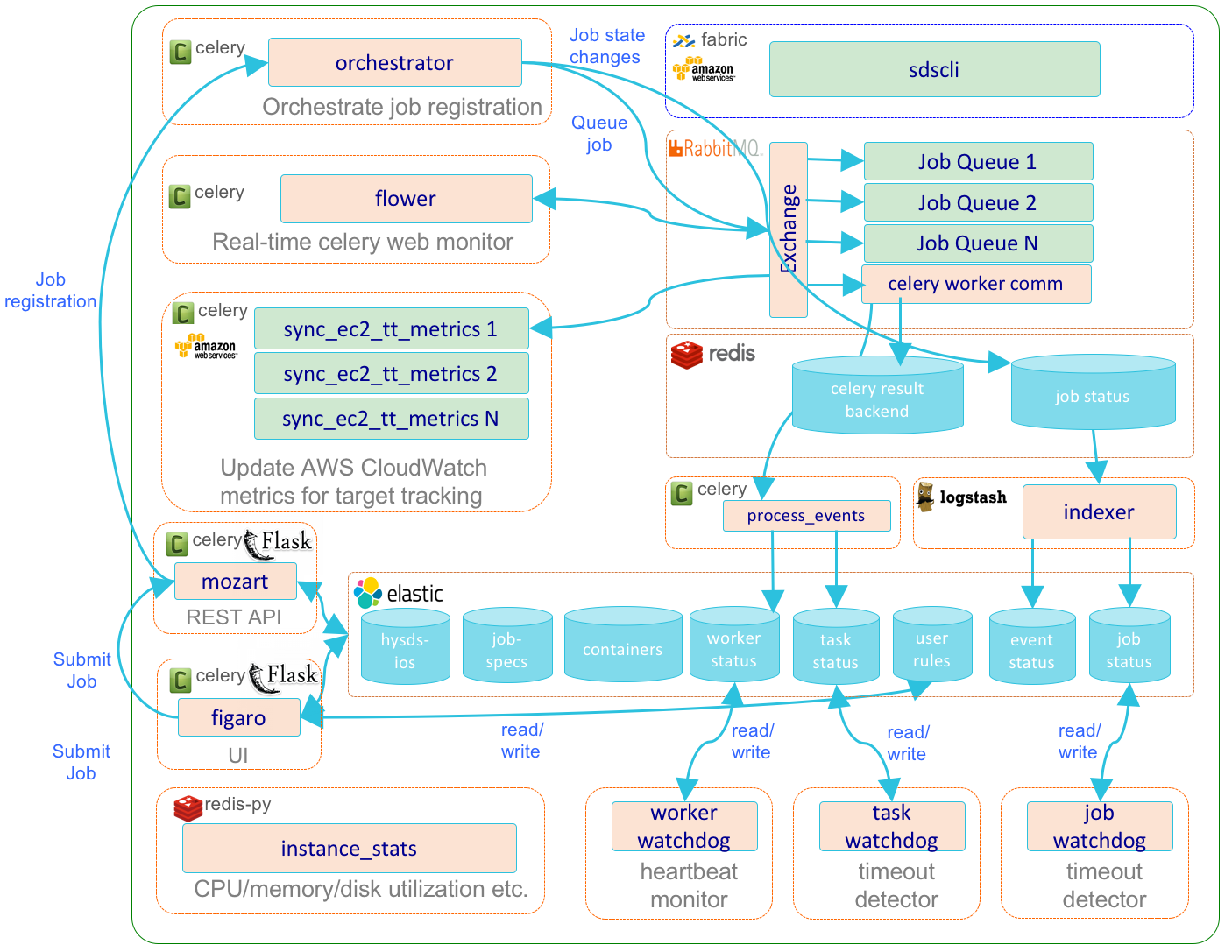
Resource Management Subsystem
The resource management subsystem (mozart) encapsulates the various 3rd party software and HySDS components related to the management of HySDS jobs/events, Celery tasks/workers, and resources used by the cluster. The diagram below depicts the dynamic interactions between these components.
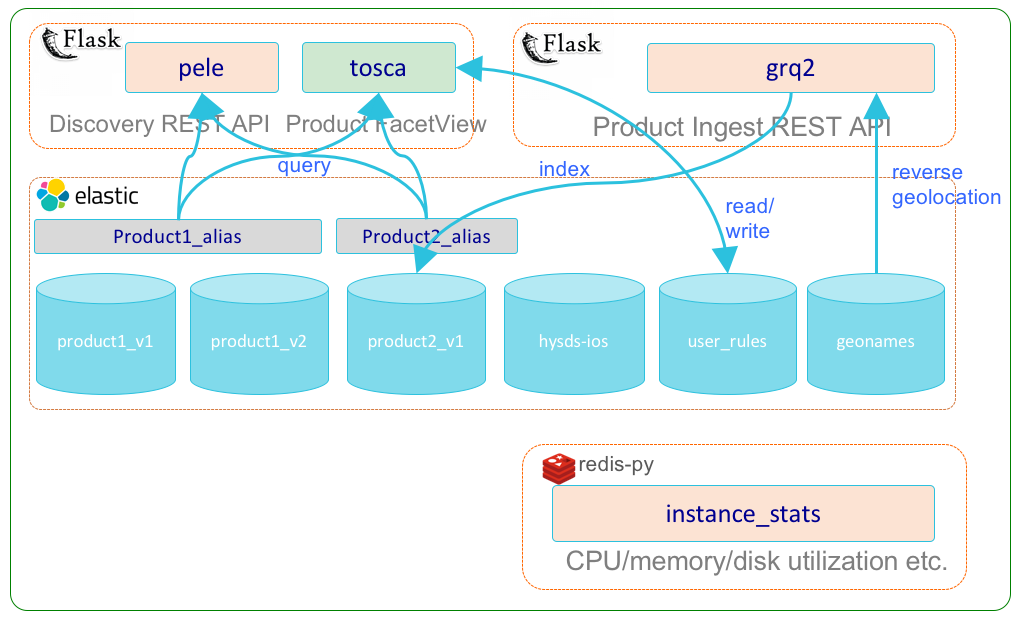
Discovery Service Subsystem
The discovery service subsystem (grq) encapsulates the various 3rd party software and HySDS components related to the management and discovery of HySDS datasets/products. The diagram below depicts the dynamic interactions between these components.
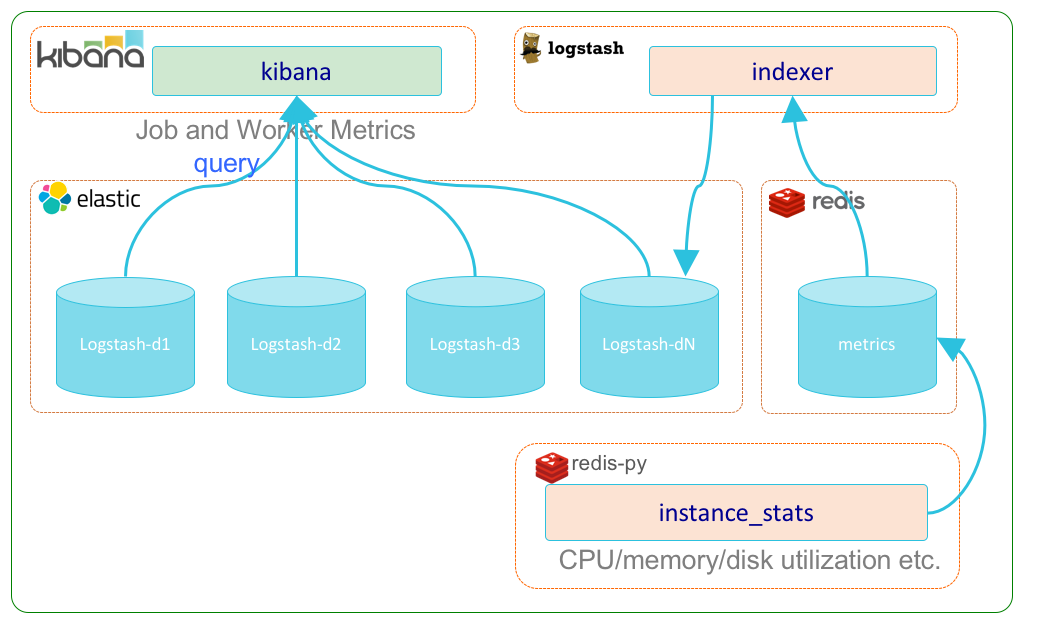
Metrics/Analytics Subsystem
The metrics/analytics subsystem (metrics) encapsulates the various 3rd party software and HySDS components related to the collection and visualization of metrics for HySDS job execution, product generation, instance resource usage (CPU, RAM, disk, I/O), and data accountability. The diagram below depicts the dynamic interactions between these components.
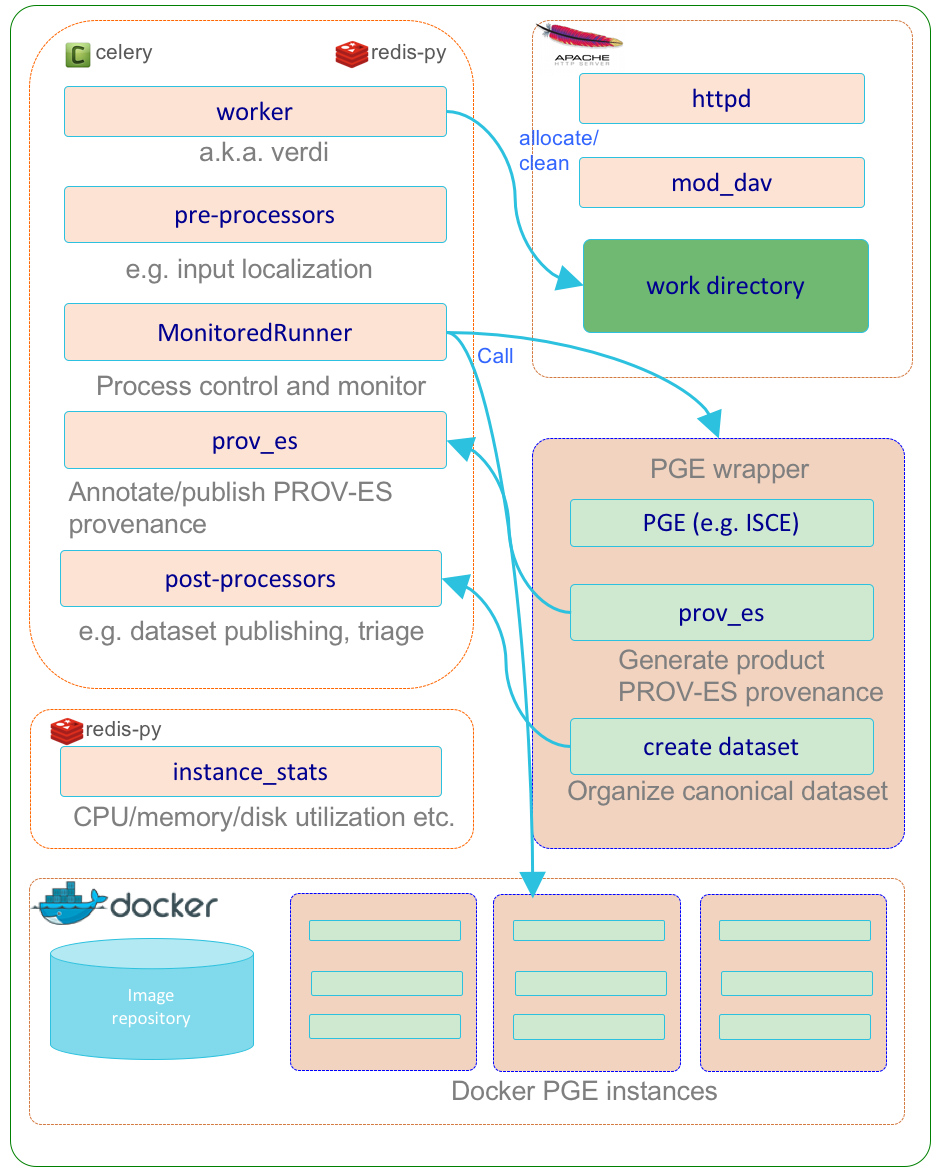
Compute Service Subsystem
The compute service subsystem (verdi) encapsulates the various 3rd party software and HySDS components related to the execution of HySDS jobs and generation of HySDS datasets/products. The diagram below depicts the dynamic interactions between these components.
Helper Subsystem
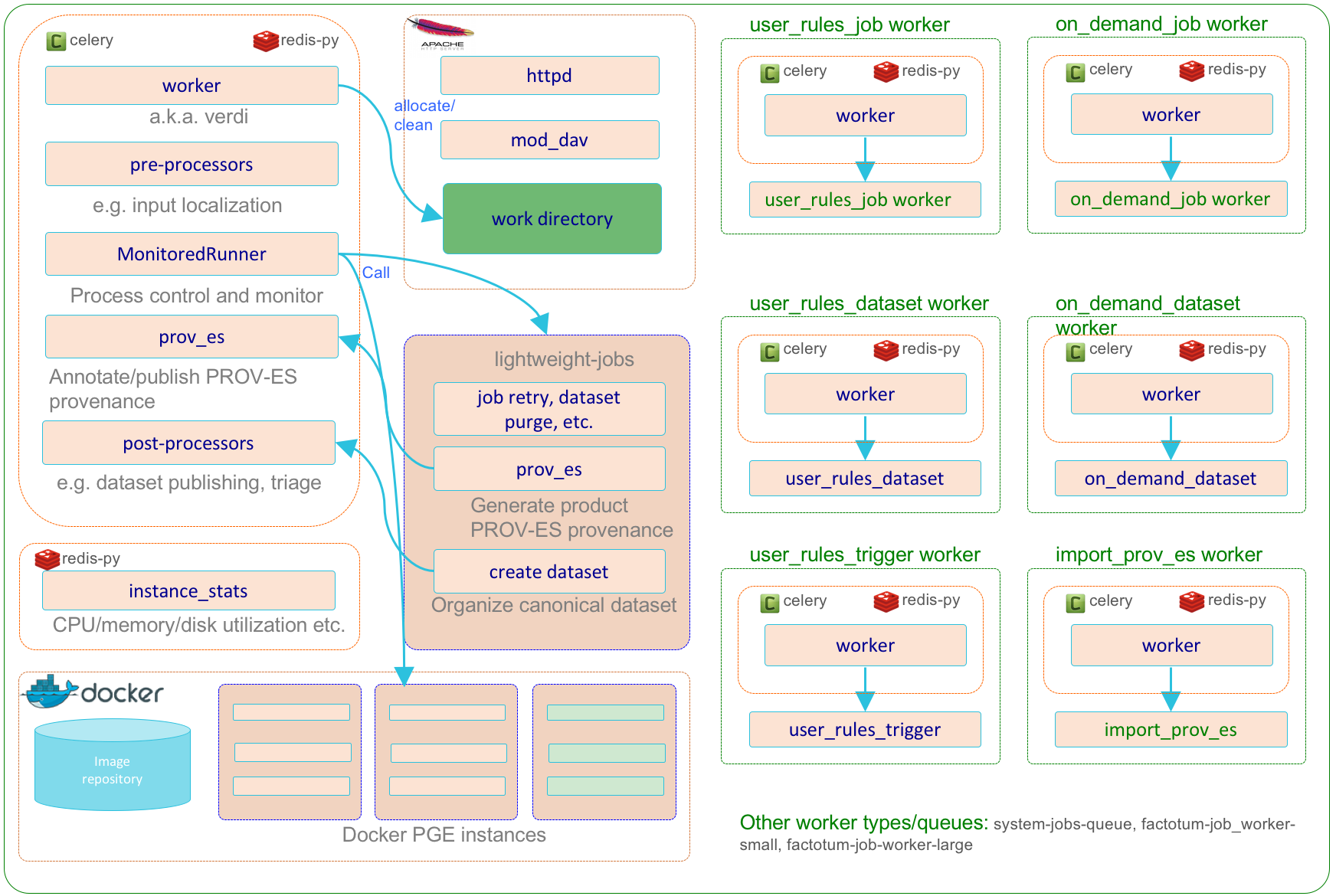
The helper subsystem (factotum) is a specialization of the compute service subsystem (verdi) that encapsulates the various 3rd party software and HySDS components related to the execution of system-based HySDS jobs and lightweight Celery tasks. The helper subsystem primarily execute jobs and tasks related to trigger rule evaluation and on-demand job submissions. Additionally, the helper subsystem can be used to run one-off Celery workers that execute HySDS jobs that does not need to scale or are throttled by external factors. The diagram below depicts the dynamic interactions between these components.
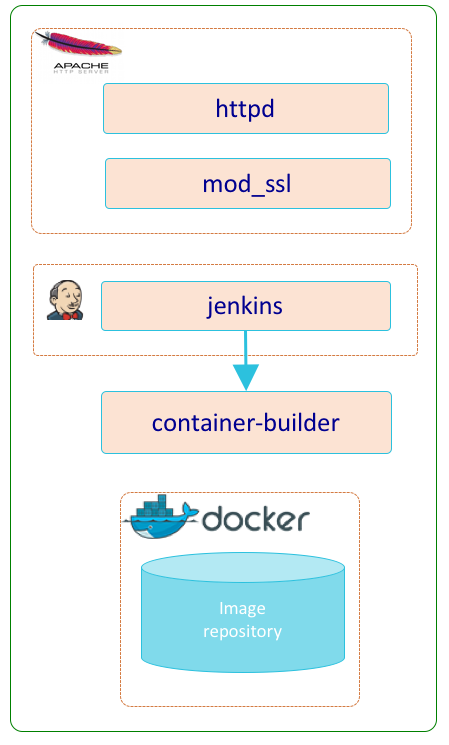
CI/CD Subsystem
The CI/CD subsystem (ci) is an optional subsystem that encapsulates the various 3rd party software and HySDS components related to the development, integration, and deployment of HySDS packages. A HySDS package is a bundled set of related HySDS job types coupled with their container image(s) packaged as an exportable/importable format. The diagram below depicts the dynamic interactions between these components.
Detailed Design
HySDS Jobs
HySDS jobs are essentially celery tasks. More specifically, they are celery tasks that encapsulate the execution of some executable within a container image. The celery task callable (hysds.job_worker.run_job) is responsible for setup, execution, and tear down of the job's work environment. Specifically, it ensures:
- there is enough free space on the root work directory (threshold defaults to 10% free)
- if there isn't, it cleans out old work directories until the threshold is met
- the job has a unique work directory to execute in
- job state is propagated to mozart
- job metrics is propagated to metrics
- pre-processing steps are executed
- container parameters such as volume mounts and UID/GID are set according to job specifications (job-spec)
- executable is run in a container
- post-processing steps are executed
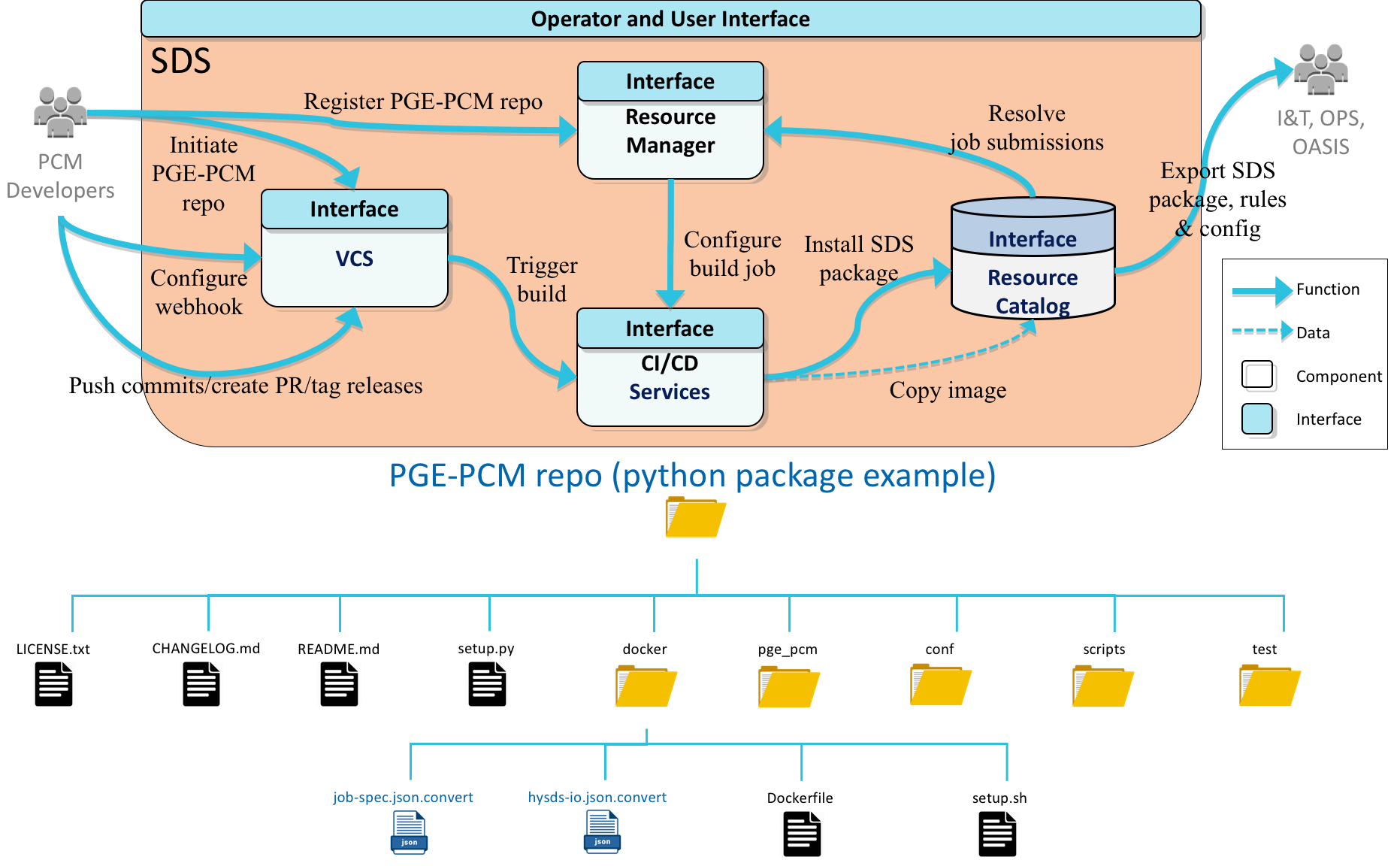
Jobs are created and integrated into a HySDS cluster by developing a job container interface as described in detail the next section. The diagram below depicts the workflow for building and integrating a job container and also shows the requisite directory structure needed for the HySDS cluster to build the container:
Requisite repo directory structure
Although the above diagram shows a python package example, the repository can contain code from any language (including shell scripts) as long as the container that is built supports those runtimes. The only requisite needed to build this repo as a job container is a top-level directory called docker which includes a Dockerfile that builds the container with all its dependencies and the job-spec and hysds-io JSON files that describe the job types.
Job Container Interface
HySDS provides capabilities to automatically manage, and continuously integrate the jobs that are in the system. In order to do that, it separates job and interface. The job-spec is used to specify a job type and the hysds-io is used to specify a job type's interface of input/output parameters. HySDS utilizes container technology to encapsulate job types and execute them on workers.
Job Specification (job-spec.json.*)
The job-spec contains the job specific information and points to the container specification that can run this job. There can be many job specifications to a single container specification. The job specification is a JSON field and requires several metadata fields in order for the job to be used by a HySDS cluster.
JSON parameters
| key | constraint | type | description |
|---|---|---|---|
| command | required | string | executable path inside container |
| params | required | array | list of param objects required to run this job (see param Object below) |
| imported_worker_files | optional | object | mapping of host file/directory into container (see imported_worker_files Object below) |
| dependency_images | optional | array | list of dependency image objects (see dependency_image Object below) |
| recommended-queues | optional | array | list of recommended queues |
| disk_usage | optional | string | minimum free disk usage required to run job specified as "\d+(GB|MB|KB)", e.g. "100GB", "20MB", "10KB" |
| soft_time_limit | optional | int | soft execution time limit in seconds; worker will send a catchable exception to task to allow for cleanup before being killed; effectively a sigterm by the worker to the job; one caveat when determining the soft time limit of your job type: also include time for verdi operations such as docker image loading (on first job), input localization, dataset publishing, triage, etc. |
| time_limit | optional | int | hard execution time limit in seconds; worker will send an uncatchable exception to the task and will force terminate it; effectively a sigkill by the worker to the job; one caveat when determining the hard time limit of your job type: make sure it's at least 60 seconds greater than the soft_time_limit otherwise job states will be orphaned in figaro |
| pre | optional | array | list of strings specifying pre-processor functions to run; behavior depends on disable_pre_builtins; more info below on Preprocessor Functions |
| disable_pre_builtins | optional | boolean | if set to true, default builtin pre-processors (currently [hysds.utils.localize_urls, hysds.utils.mark_localized_datasets, hysds.utils.validate_checksum_files]) are disabled and would need to be specified in pre to run; if not specified or set to false, list of pre-processors specified by pre is appended after the default builtins |
| post | optional | array | list of strings specifying post-processor functions to run; behavior depends on disable_post_builtins; more info below on Postprocessor Functions |
| disable_post_builtins | optional | boolean | if set to true, default builtin post-processors (currently [hysds.utils.publish_datasets]) are disabled and would need to be specified in post to run; if not specified or set to false, list of post-processors specified by post is appended after the default builtins |
param Object
| key | constraint | type | description |
|---|---|---|---|
| name | required | string | parameter name |
| destination | required | string |
|
imported_worker_files Object
| key | key type | value | value type |
|---|---|---|---|
| path to file or directory on host | string | path to file or directory in container | string |
| path to file or directory on host | string | one item list of path to file or directory in container | array |
| path to file or directory on host | string | two item list of path to file or directory in container and mount mode: ro for read-only and rw for read-write (default is ro) | array |
Syntax
{
"command": "string",
"recommended-queues": [ "string" ],
"disk_usage":"\d+(GB|MB|KB)",
"imported_worker_files": {
"string": "string",
"string": [ "string" ],
"string": [ "string", "ro" | "rw" ]
},
"dependency_images": [
{
"container_image_name": "string",
"container_image_url": "string" | null,
"container_mappings": {
"string": "string",
"string": [ "string" ],
"string": [ "string", "ro" | "rw" ]
}
}
],
"soft_time_limit": int,
"time_limit": int,
"disable_pre_builtins": true | false,
"pre": [ "string" ],
"disable_post_builtins": true | false,
"post": [ "string" ],
"params": [
{
"name": "string",
"destination": "positional" | "localize" | "context"
}
]
}
Example
{
"command": "/home/ops/verdi/ops/hello_world/run_hello_world.sh",
"recommended-queues": [ "factotum-job_worker-small" ],
"disk_usage":"10MB",
"imported_worker_files": {
"$HOME/.netrc": ["/home/ops/.netrc"],
"$HOME/.aws": ["/home/ops/.aws", "ro"],
"$HOME/ariamh/conf/settings.conf": "/home/ops/ariamh/conf/settings.conf"
},
"dependency_images": [
{
"container_image_name": "aria/isce_giant:latest",
"container_image_url": "s3://s3-us-west-2.amazonaws.com/my-hysds-code-bucket/aria-isce_giant-latest.tar.gz",
"container_mappings": {
"$HOME/.netrc": ["/root/.netrc"],
"$HOME/.aws": ["/root/.aws", "ro"]
}
}
],
"soft_time_limit": 900,
"time_limit": 960,
"disable_pre_builtins": false,
"pre": [ "some.python.preprocessor.function" ],
"disable_post_builtins": false,
"post": [ "hysds.utils.triage" ],
"params": [
{
"name": "localize_url",
"destination": "localize"
},
{
"name": "file",
"destination": "positional"
},
{
"name": "prod_name",
"destination": "context"
}
]
}
Preprocessor and Postprocessor Functions
HySDS provides a plugin facility to hook in arbitrary function calls before (pre-processor functions) and after (post-processor functions) the execution of a docker command in a job. HySDS provides builtin pre-processor functions that runs by default for each job (hysds.utils.localize_urls, hysds.utils.mark_localized_datasets, hysds.utils.validate_checksum_files) to localize any inputs specified in the job payload. It also provides builtin post-processor functions that runs by default as well (hysds.utils.publish_datasets) to search the work directory for any HySDS datasets to publish.
Function Definition
Pre-processor and post-processor functions are required to take 2 arguments: the job dict and context dict. These functions can do what they need using information stored in the job and context dicts. The function must return a boolean result, either True or False. In the case of pre-processor functions, the docker command for the job is only executed if all pre-processor functions return a True. In the case of post-processor functions, any that return a False will be logged but the job will not be considered a failure. In both cases, if the function raises an exception, then the job is a failure.
Pre-processor Functions
By default, job-specs have an implicit pre-processor function defined, hysds.utils.localize_urls. Thus the following job-spec:
{
"command":"/home/ops/ariamh/ariaml/extractFeatures.sh",
"disk_usage":"1GB",
"imported_worker_files": {
"/home/ops/.netrc": "/home/ops/.netrc",
"/home/ops/.aws": "/home/ops/.aws",
"/home/ops/ariamh/conf/settings.conf": "/home/ops/ariamh/conf/settings.conf"
},
"params" : [
{
"name": "uri",
"destination": "positional"
},
{
"name": "uri",
"destination": "localize"
}
]
}
implicitly provides the "pre" parameter as follows:
{
"command":"/home/ops/ariamh/ariaml/extractFeatures.sh",
"disk_usage":"1GB",
"imported_worker_files": {
"/home/ops/.netrc": "/home/ops/.netrc",
"/home/ops/.aws": "/home/ops/.aws",
"/home/ops/ariamh/conf/settings.conf": "/home/ops/ariamh/conf/settings.conf"
},
"pre": [ "hysds.utils.localize_urls" ],
"params" : [
{
"name": "uri",
"destination": "positional"
},
{
"name": "uri",
"destination": "localize"
}
]
}
To disable this builtin feature, specify the "disable_pre_builtins" parameter as follows:
{
"command":"/home/ops/ariamh/ariaml/extractFeatures.sh",
"disk_usage":"1GB",
"imported_worker_files": {
"/home/ops/.netrc": "/home/ops/.netrc",
"/home/ops/.aws": "/home/ops/.aws",
"/home/ops/ariamh/conf/settings.conf": "/home/ops/ariamh/conf/settings.conf"
},
"disable_pre_builtins": true,
"params" : [
{
"name": "uri",
"destination": "positional"
},
{
"name": "uri",
"destination": "localize"
}
]
}
In this case, even though there exists a parameter "uri" with destination "localize", the hysds.utils.localize_urls pre-processor will not run. To specify additional pre-processor functions, define them using the "pre" parameter like so:
{
"command":"/home/ops/ariamh/ariaml/extractFeatures.sh",
"disk_usage":"1GB",
"imported_worker_files": {
"/home/ops/.netrc": "/home/ops/.netrc",
"/home/ops/.aws": "/home/ops/.aws",
"/home/ops/ariamh/conf/settings.conf": "/home/ops/ariamh/conf/settings.conf"
},
"pre": [ "my.custom.preprocessor_function" ],
"params" : [
{
"name": "uri",
"destination": "positional"
},
{
"name": "uri",
"destination": "localize"
}
]
}
Because the default builtin pre-processor functions aren't disabled, the effect of the above job-spec is this:
{
"command":"/home/ops/ariamh/ariaml/extractFeatures.sh",
"disk_usage":"1GB",
"imported_worker_files": {
"/home/ops/.netrc": "/home/ops/.netrc",
"/home/ops/.aws": "/home/ops/.aws",
"/home/ops/ariamh/conf/settings.conf": "/home/ops/ariamh/conf/settings.conf"
},
"disable_pre_builtins": true,
"pre": [ "hysds.utils.localize_urls", "my.custom.preprocessor_function" ],
"params" : [
{
"name": "uri",
"destination": "positional"
},
{
"name": "uri",
"destination": "localize"
}
]
}
Post-processor Functions
By default, job-specs have an implicit post-processor function defined, hysds.utils.publish_datasets. Thus the following job-spec:
{
"command":"/home/ops/ariamh/ariaml/extractFeatures.sh",
"disk_usage":"1GB",
"imported_worker_files": {
"/home/ops/.netrc": "/home/ops/.netrc",
"/home/ops/.aws": "/home/ops/.aws",
"/home/ops/ariamh/conf/settings.conf": "/home/ops/ariamh/conf/settings.conf"
},
"params" : [
{
"name": "uri",
"destination": "positional"
},
{
"name": "uri",
"destination": "localize"
}
]
}
implicitly provides the "post" parameter as follows:
{
"command":"/home/ops/ariamh/ariaml/extractFeatures.sh",
"disk_usage":"1GB",
"imported_worker_files": {
"/home/ops/.netrc": "/home/ops/.netrc",
"/home/ops/.aws": "/home/ops/.aws",
"/home/ops/ariamh/conf/settings.conf": "/home/ops/ariamh/conf/settings.conf"
},
"post": [ "hysds.utils.publish_datasets" ],
"params" : [
{
"name": "uri",
"destination": "positional"
},
{
"name": "uri",
"destination": "localize"
}
]
}
To disable this builtin feature, specify the "disable_post_builtins" parameter as follows:
{
"command":"/home/ops/ariamh/ariaml/extractFeatures.sh",
"disk_usage":"1GB",
"imported_worker_files": {
"/home/ops/.netrc": "/home/ops/.netrc",
"/home/ops/.aws": "/home/ops/.aws",
"/home/ops/ariamh/conf/settings.conf": "/home/ops/ariamh/conf/settings.conf"
},
"disable_post_builtins": true,
"params" : [
{
"name": "uri",
"destination": "positional"
},
{
"name": "uri",
"destination": "localize"
}
]
}
In this case, even though the docker command may create a HySDS dataset in the job's working directory, the hysds.utils.publish_urls post-processor will not run. To specify additional post-processor functions, define them using the "post" parameter like so:
{
"command":"/home/ops/ariamh/ariaml/extractFeatures.sh",
"disk_usage":"1GB",
"imported_worker_files": {
"/home/ops/.netrc": "/home/ops/.netrc",
"/home/ops/.aws": "/home/ops/.aws",
"/home/ops/ariamh/conf/settings.conf": "/home/ops/ariamh/conf/settings.conf"
},
"post": [ "my.custom.postprocessor_function" ],
"params" : [
{
"name": "uri",
"destination": "positional"
},
{
"name": "uri",
"destination": "localize"
}
]
}
Because the default builtin post-processor functions aren't disabled, the effect of the above job-spec is this:
{
"command":"/home/ops/ariamh/ariaml/extractFeatures.sh",
"disk_usage":"1GB",
"imported_worker_files": {
"/home/ops/.netrc": "/home/ops/.netrc",
"/home/ops/.aws": "/home/ops/.aws",
"/home/ops/ariamh/conf/settings.conf": "/home/ops/ariamh/conf/settings.conf"
},
"disable_pre_builtins": true,
"post": [ "hysds.utils.publish_datasets", "my.custom.preprocessor_function" ],
"params" : [
{
"name": "uri",
"destination": "positional"
},
{
"name": "uri",
"destination": "localize"
}
]
}
Triage
HySDS provides an additional builtin post-processor function for providing triage of failed jobs: hysds.utils.triage. It is not enabled by default and must be explicitly set:
{
"command":"/home/ops/ariamh/ariaml/extractFeatures.sh",
"disk_usage":"1GB",
"imported_worker_files": {
"/home/ops/.netrc": "/home/ops/.netrc",
"/home/ops/.aws": "/home/ops/.aws",
"/home/ops/ariamh/conf/settings.conf": "/home/ops/ariamh/conf/settings.conf"
},
"post": [ "hysds.utils.triage" ],
"params" : [
{
"name": "uri",
"destination": "positional"
},
{
"name": "uri",
"destination": "localize"
}
]
}
The above job-spec results in 2 post-processing functions being called after the docker command for the job executes: hysds.utils.publish_datasets and hysds.utils.triage. In both cases, the functions perform the proper checks up-front before continuing on with their functionality. For example, hysds.utils.publish_datasets checks the exit code of the docker command by inspecting the job dict and only proceeds with searching for HySDS datasets if the exit code was 0. For hysds.utils.triage, the function checks that the exit code was not a 0 and continues with the creation and publishing of the triage dataset if so. If the job was triaged, there will be a _triaged.json file left in the job's work directory which contains the JSON result returned by the GRQ ingest REST call.
What gets triaged?
By default, all _* and *.log files in the root of the job work directory are triaged.
Can we triage other files as well?
The behavior of the triage function can be modified by updating certain fields in the _context.json of the job's work directory. Within the docker command that is called, the process can open up the _context.json and add a top-level parameter named _triage_additional_globs as a list of glob patterns that will be triaged in addition to the default files. For example, a python script that is called as a docker command for the job can add these files for triage:
with open(ctx_file) as f:
ctx = json.load(f)
ctx['_triage_additional_globs'] = [ 'S1-IFG*', 'AOI_*', 'celeryconfig.py', 'datasets.json' ]
with open(ctx_file, 'w') as f:
json.dump(ctx, f, sort_keys=True, indent=2)
Triage is enabled in the job-spec. Can we disable triage at runtime?
Yes. Similar to the addition of other files to triage, you can add a top-level parameter named _triage_disabled to disable triage:
with open(ctx_file) as f:
ctx = json.load(f)
ctx['_triage_additional_globs'] = [ 'S1-IFG*', 'AOI_*', 'celeryconfig.py', 'datasets.json' ]
if some_condition:
ctx['_triage_disabled'] = True
with open(ctx_file, 'w') as f:
json.dump(ctx, f, sort_keys=True, indent=2)
HySDS IO Specification (hysds-io.json.*)
The hysds-io file is designed to create a wiring to the specified params defined in the job-spec. It specifies the from-where as opposed to the to-where, which is specified by the job-spec. It has several fields defined in its JSON syntax.
JSON Parameters
| key | constraint | type | description |
|---|---|---|---|
| component | optional | string | component web interface to display this job type in (tosca or figaro); defaults to tosca |
| label | optional | string | label to be used when this job type is displayed in web interfaces (tosca and figaro); otherwise it will show an automatically generated label based on the string after the "hysds.io.json." of the hysds-io file |
| submission_type | optional | string | specifies if the job should be submitted once per product in query or once per job submission; iteration for a submission of the job for each result in the query or individual for a single submission; defaults to iteration |
| enable_dedup | optional | boolean | set to true to enable job deduplication; false to disable; defaults to true |
| action-type | optional | string | action type to expose job as; on-demand, trigger, or both; defaults to both |
| allowed_accounts | optional | array | list of strings specifying account user IDs allowed to run this job type from the web interfaces (tosca and figaro); if not defined, ops account is the only account that can access this job type; if _all is specified in the list, then all users will be able to access this job type |
| params | required | array | list of matching param objects from job-spec required to run this job (see params Object below) |
A Note on submission_type: iteration and individual
Iteration jobs submit one job for each of the faceted results in the selected set (tosca or figaro). This means that each parameter using "dataset_jpath:" (see below) submits a unique job where the value of that parameter corresponds to an individual product from the faceted set.
Individual jobs simply submit one job, no matter how many products are faceted. If "dataset_jpath:" is specified, the corresponding values are aggregated into a list, and that becomes the value for the parameter below.
params Object
| key | constraint | type | description |
|---|---|---|---|
| name | required | string | parameter name; should match corresponding parameter name in job-spec |
| from | required | string | value for hard-coded parameter value, submitter for user submitted value, dataset_jpath:<jpath> to extract value from ElasticSearch result using a JPath-like notation (see from Specification below) |
| value | required if from is set to value | string | hard-coded parameter value |
| type | optional | string | possible values: text, number, datetime, date, boolean, enum, email, textarea, region, container_version, jobspec_version, hysdsio_version (see Valid Param Types below) |
| default | optional | string | default value to use (must be string even if it's a number) |
| optional | optional | boolean | parameter is optional and can be left blank |
| placeholder | optional | string | value to use as a hint when displaying the form input |
| enumerables | required if type is enum | array | list of string values to enumerate via a dropdown list in the form input |
| lambda | optional | string | a lambda function to process the value during submission |
| version_regex | required if type is container_version, jobspec_version or hysdsio_version | string | regex to use to filter on front component of respective container, job-spec or hysds-io ID; e.g. if type=jobspec_version, version_regex=job-time-series and list of all job-specs IDs in the system is ["job-time-series:release-20171103", "job-hello_world:release-20171103", job-time-series:master"], the list will be filtered down to those whose ID matched the regex job-time-series in the front component (string before the first ":"); in this example the resulting filtered set of release tags/branches will be listed as ["release-20171103", "master"] in a dropdown box; similar to type=enum and enumerable set to all release tags for a certain job type |
from Specification
| value | description |
|---|---|
| value | hard-coded parameter value found in value field in hysds-io.json |
| submitter | comes from operator or submitter of job; usually displayed as input boxes on the web UI forms for tosca and figaro |
| dataset_jpath:<jpath> | comes from an ElasticSearch result, e.g. in tosca, the iterated dataset's metadata supplies this field; metadata is mined from the metadata found at the "." separated path to a given metadata field. i.e. metadata.field1 pulls from the metadata field's field1 value; the top level of the jpath is the top of the ES entry. i.e. dataset_jpath:_id is a valid dataset jpath to pull out ES's ID |
Valid Param Types
| value | description |
|---|---|
| text | a text string, will be kept as text |
| number | a real number |
| date | a date in ISO8601 format: YYYY-MM-DD; will be treated as a "text" field for passing into the container |
| datetime | a date with time in ISO8601 format: YYYY-MM-DDTHH:mm:SS.SSS; will be treated as a "text" field |
| boolean | true or false in a drop down |
| enum | one of a set of options in a drop down; must specify enumerables field to specify the list of possible options; these will be "text" types in the enumerables set |
| an e-mail address, will be treated as "text" | |
| textarea | same as text, but displayed larger with the textarea HTML tag |
| region | auto-populated from the facet view leaflet tool |
| container_version | a version of an existing container registered in the Mozart API; must define "version_regex" field |
| jobspec_version | a version of an existing job-spec registered in the Mozart API; must define "version_regex" field |
| hysdsio_version | a version of an existing hysds-io registered in the Mozart API; must define "version_regex" field |
A note on type conversion and default values
Due to a limitation in ElasticSearch mappings, hysds-io "default_values" should be represented as JSON strings. This parallels the input from the user which will come through as a string. Internally the system will convert the value into its "type" within python. This happens just before the call to a "lambda" function and thus the user can expect proper types inside the lambda function.
Certain types default to python strings. these types include: "container_version", "hysdsio_version", "jobspec_version", "date", "datetime", "text", "string", "textbox", "enum", and "email".
All numbers are converted into floats due to the nature of JSON which does not distinguish between integer and float values. If a python integer is needed by the job, supply a lambda like so "lambda x: int(x)".
Syntax
{
"label": "string",
"submission_type": "individual" | "iteration",
"allowed_accounts": [ "string" ],
"params": [
{
"name": "string",
"from": "value" | "submitter" | "dataset_jpath:<path>",
"type": "string",
"default": "string",
"placeholder": "string",
"enumerables": [ "string" ],
"lambda": "string",
"value": "string",
"version_regex": "string"
}
]
}
Example
{
"label": "Hello World",
"submission_type": "iteration",
"allowed_accounts": [ "ops" ],
"params": [
{
"name": "localize_url",
"from": "dataset_jpath:_source",
"lambda": "lambda ds: \"%s/%s\" % ((filter(lambda x: x.startswith('s3://'), ds['urls']) or ds['urls'])[0], ds['metadata']['data_product_name'])"
},
{
"name": "file",
"from": "dataset_jpath:_source.metadata.data_product_name"
},
{
"name": "prod_name",
"from": "dataset_jpath:_source.metadata.prod_name"
}
]
}
Dataset Specification
In order to for HySDS to recognize a dataset, the dataset must follow certain conventions.
Dataset ID
Each product should have a dataset ID. This name is used to determine the type of the dataset and name all the important files for the dataset. A dataset ID is matched against entries found in the datasets.json file to determine its type.
In this example, we shall use the dataset ID dumby-product-20170101T000000Z-3lx0a.
Directory
Any directory containing the below JSON files and found within the working directory supplied to the job is considered a dataset. Thus this directory must be named with the dataset's ID (see above):
$ pwd /data/work/example_work_dir/dumby-product-20170101T000000Z-3lx0a $ ls dumby-product-20170101T000000Z-3lx0a.dataset.json dumby-product-20170101T000000Z-3lx0a.met.json dumby-product-20170101T000000Z-3lx0a.prov_es.json dumby-product-20170101T000000Z-3lx0a.h5 pge_output_2.h5 errors.txt other_metadata.xml
Note that any other PGE data files should be placed in the <Dataset ID> directory, as the whole directory is the dataset.
HySDS dataset and metadata JSON files
dataset JSON file
A product must produce a <Dataset ID>.dataset.json in the <Dataset ID> directory. This file contains JSON formatted metadata representing the cataloged dataset metadata:
$ cat dumby-product-20170101T000000Z-3lx0a.dataset.json
{
"version": "v1.0",
"label": "dumby product for 2017-01-01T00:00:00Z",
"location": {
"type": "polygon",
"coordinates": [
[
[-122.9059682940358,40.47090915967475],
[-121.6679748715316,37.84406528996276],
[-120.7310161872557,38.28728069813177],
[-121.7043611684245,39.94137004454238],
[-121.9536916840953,40.67097860759095],
[-122.3100379696548,40.7267890636145],
[-122.7640648263371,40.5457010812299],
[-122.9059682940358,40.47090915967475]
]
]
},
"starttime": "2017-01-01T00:00:00",
"endtime": "2017-01-01T00:05:00"
}
The required fields are:
- version
The optional fields are:
- label
- location (in GeoJSON format)
- starttime
- endtime
metadata JSON file
In addition, other metadata data can be added to a <Dataset ID>.met.json in the <Dataset ID> directory. As long as the file conforms to the JSON format, the dataset developer has free reign on what goes into this file:
$ cat dumby-product-20170101T000000Z-3lx0a.met.json
{
"startingRange": 800026.4431219272,
"sensor": "SAR-C Sentinel1",
"esd_threshold": 0.85,
"tiles": true,
"reference": true,
"trackNumber": 144,
"lookDirection": "right",
"beamMode": "IW",
"direction": "descending",
"inputFile": "sentinel.ini",
"polarization": "VV",
"imageCorners": {
"maxLon": -117.56055555555555,
"minLon": -119.06166666666667,
"minLat": 35.92333333333333,
"maxLat": 37.766666666666666
},
...
"orbitRepeat": 175
}
PROV-ES JSON file (optional)
If the developer has instrumented the PGE to use the PROV-ES (Provenence for Earth Science) service, a <Dataset ID>.prov_es.json file must exist in the <Dataset ID> directory containing the PROV-ES JSON:
$ cat dumby-product-20170101T000000Z-3lx0a.prov_es.json
{
"wasAssociatedWith": {
"hysds:a379e0e0-32ac-59b7-89b8-80045bad1ba0": {
"prov:agent": "hysds:47bce551-6fac-5bb5-adcc-61918c608f96",
"prov:activity": "hysds:8e5a0556-99b5-56a9-ae0b-e1de3f5fe216"
}
},
"used": {
"hysds:d04421e2-9847-5073-9f45-1b3e0b011cd8": {
"prov:role": "input",
"prov:time": "2015-09-13T14:18:39.037508+00:00",
"prov:entity": "hysds:e08c901b-2f98-514a-98cc-76f5a124ccd6",
"prov:activity": "hysds:8e5a0556-99b5-56a9-ae0b-e1de3f5fe216"
}
},
"agent": {
"eos:JPL": {
"prov:type": {
"type": "prov:QualifiedName",
"$": "prov:Organization"
},
"prov:label": "Jet Propulsion Laboratory"
},
"hysds:47bce551-6fac-5bb5-adcc-61918c608f96": {
"hysds:host": "x.x.x.x",
"prov:label": "hysds:pge_wrapper/x.x.x.x/20202/2015-09-13T14:18:39.866510",
"prov:type": {
"type": "prov:QualifiedName",
"$": "prov:SoftwareAgent"
},
"hysds:pid": "20202"
},
...
"prefix": {
"info": "http://info-uri.info/",
"bibo": "http://purl.org/ontology/bibo/",
"hysds": "http://hysds.jpl.nasa.gov/hysds/0.1#",
"eos": "http://nasa.gov/eos.owl#",
"gcis": "http://data.globalchange.gov/gcis.owl#",
"dcterms": "http://purl.org/dc/terms/"
},
"activity": {
"hysds:8e5a0556-99b5-56a9-ae0b-e1de3f5fe216": {
"eos:runtimeContext": "hysds:runtimeContext-ariamh-trigger_pietro_use_case_triggersf",
"hysds:job_url": "http://x.x.x.x:8085/jobs/2015/09/13/create_interferogram-CSKS2_RAW_HI_04_HH_RA_20110811135636_20110811135643.interferogram.json_10-20150913T072449.918362Z",
"prov:startTime": "2015-09-13T13:36:29.047370+00:00",
"prov:type": "hysds:create_interferogram",
"hysds:mozart_url": "https://x.x.x.x:8888",
"eos:usesSoftware": [
"eos:ISCE-2.0.0_201410"
],
"hysds:job_type": "create_interferogram",
"hysds:job_id": "create_interferogram-CSKS2_RAW_HI_04_HH_RA_20110811135636_20110811135643.interferogram.json_10-20150913T072449.918362Z",
"prov:endTime": "2015-09-13T14:18:39.835057+00:00",
"prov:label": "create_interferogram-2015-09-13T14:18:39.037508Z"
}
},
"wasGeneratedBy": {
"hysds:bc3f219a-4002-57d8-8151-9fe3b7776d40": {
"prov:role": "output",
"prov:time": "2015-09-13T14:18:39.037508+00:00",
"prov:entity": "hysds:6534285c-01f0-5070-8294-a3847b048d5b",
"prov:activity": "hysds:8e5a0556-99b5-56a9-ae0b-e1de3f5fe216"
}
}
}
Dataset Configuration
Once a PGE is configured to generate a dataset according to the HySDS dataset specification above, users will have to configure HySDS to recognize the new dataset type so that they get ingested properly into the system.
datasets JSON config
The datasets.json configuration file is centrally stored at some location (usually on the mozart instance in the ops user's ~/.sds/files directory) and is published to a repository where instantiating workers can pull from. At job execution time, the datasets.json configuration file is placed in the work directory of the job for the dataset recognition step. The format for datasets.json is:
{
"datasets": [
{
"ipath": "hysds::data/dumby-product",
"version": "v0.1",
"level": "l0",
"type": "dumby-data",
"id_template": "${id}",
"match_pattern": "/(?P<id>dumby-product-\\d+)$",
"alt_match_pattern": null,
"extractor": null,
"publish": {
"s3-profile-name": "<aws creds profile name>",
"location": "<S3 url to S3 bucket>/products/dumby/${id}",
"urls": [
"<HTTP url to S3 bucket>/products/dumby/${id}",
"<S3 url to S3 bucket>/products/dumby/${id}"
]
},
"browse": {
"s3-profile-name": "<aws creds profile name>",
"location": "<S3 url to S3 bucket>/browse/dumby/${id}",
"urls": [
"<HTTP URL to S3 bucket>/browse/dumby/${id}",
"<S3 URL to S3 bucket>/browse/dumby/${id}"
]
}
}
]
}
At its core the file is a list of data set objects associated with the key "datasets". Each data set object specifies the following properties:
- ipath: a namespaced name for the product type
- version: a version string for the product
- level: a level (L0, L1, L2, L3 ...) string for the product
- type: type name for the data product
- id_template: a template to build the id for the product from capture groups from in the match_pattern (#6)
- match_pattern: regular expression to match product name strings (from sub-directories in PGE working directory). Should always start with a '/'.
- alt_match_pattern: alternate pattern to #6 (usually just null)
- extractor: path to executable binary or script to run further metadata extraction on the dataset
- publish: an object specifying how to publish this product composed of the following fields
- s3-profile-name: AWS profile to use as defined in ~/.aws.
- location: Location for data-store location of final archived product using Osaka prefix
- urls: List of urls pointing to the product once archived. Can include http access, s3 access or any other URL type
- browse: an object with the same attributes as publish (#9) but handling the publishing of browse-images
For example, the datasets.json file below shows the configuration for our dumby dataset (dumby-product-20170101T000000Z-3lx0a):
{
"datasets": [
{
"ipath": "hysds::data/dumby-product",
"match_pattern": "/(?P<id>dumby-product-\\d+)$",
"alt_match_pattern": null,
"extractor": null,
"level": "l0",
"type": "dumby-data",
"publish": {
"s3-profile-name": "default",
"location": "s3://s3-us-west-2.amazonaws.com:80/my_bucket/products/dumby/${version}/${id}",
"urls": [
"http://my_bucket.s3-website-us-west-2.amazonaws.com/products/dumby/${version}/${id}",
"s3://s3-us-west-2.amazonaws.com:80/my_bucket/products/dumby/${version}/${id}"
]
},
"browse": {
"s3-profile-name": "default",
"location": "s3://s3-us-west-2.amazonaws.com:80/my_bucket/browse/dumby/${version}/${id}",
"urls": [
"http://my_bucket.s3-website-us-west-2.amazonaws.com/browse/dumby/${version}/${id}",
"s3://s3-us-west-2.amazonaws.com:80/my_bucket/browse/dumby/${version}/${id}"
]
}
}
]
}
Job Execution
The following sequence diagram presented in a previous section describes the detailed interactions between the various subsystems and components during job execution.
The table below briefly describes the objects/actors that play a role in job execution:
| object/actor | subsystem | component | description |
|---|---|---|---|
| Submitter |
| hysds | Job submitter could be:
|
| Orchestrator | resource management | hysds | Receives job submission requests, resolves job type and parameters, creates a validated job payload, and routes it to the destination queue |
| Watchdog | resource management | hysds | Daemon processes that monitor events streaming into the system to detect and mitigate anomalous events |
| Resource Catalog | resource management | mozart | Catalog interface for management, tracking, and monitoring of registered job types, job containers, job states, task states, workers, and events |
| Job Container Store | resource management | object store | Highly scalable object store for fast access to file objects related to jobs and worker configuration |
| Queue | resource management | rabbitmq | High performance message broker/queueing system |
| Worker | worker | hysds | Worker that pulls a job from the queue and executes it |
| Helper | helper | hysds | Specialized worker that runs system jobs, e.g. trigger rule evaluations |
| Dataset Catalog | discovery | grq2 | Catalog interface for management of datasets and products |
| Dataset Store | discovery | object store | Highly scalable object store for fast access to file objects related to datasets and products |
| Provenance | resource management | prov_es | PROV-ES (W3C PROV) endpoint for recording provenance traces |
| External Source | external | external | External data source, e.g. USGS DEMs, LandSat 8 Dataset in AWS |
Assuming a job type was registered into the HySDS cluster, the following sequence of events occurs:
- Submitter submits a job to the Orchestrator.
- Orchestrator parses the job request and resolves the container image mapped to the job by querying the Resource Catalog.
- Orchestrator constructs the validated job payload.
- Orchestrator enqueues the job payload to the requested Queue.
- Orchestrator signals Resource Catalog to set job state to job-queued.
- Worker listening to the Queue pulls the job payload and parses it for job information.
- Worker signals Resource Catalog to set job state to job-started.
- Watchdog monitors worker heartbeats and signals Resource Catalog to set job state to job-offline if it detects Worker is offline.
- Worker pulls the container image for the job from the Job Container Store and loads it.
- Worker proceeds with all configured preprocessors.
- Input files configured for the job are localized to the job's working directory.
- Input files with accompanying checksum files are validated.
- Localized HySDS datasets are tagged as inputs to prevent republishing.
- Worker runs the job via the job container and localize inputs.
- Worker proceeds with all configured postprocessors.
- Worker scours work directory for HySDS datasets and publishes them.
- Worker copies HySDS dataset to Dataset Store.
- Worker indexes HySDS dataset metadata into Dataset Catalog.
- If triage was configured, Worker creates a triage worker.
- Worker scours work directory for HySDS datasets and publishes them.
- Worker submits final job state to Resource Catalog: job-completed or job-failed.
- Worker submits an evaluator task to the Queue before it exits its job execution.
- Helper listening to the Queue pulls the evaluator task.
- Helper determines if any jobs can be triggered by querying the Resource Catalog and Dataset Catalog.
- Helper submits jobs to the Orchestrator if any trigger rules matched from evaluation.
Workflow
HySDS supports workflows of 2 types: implicit and explicit. This section provides an overview of these core workflow capabilities built into HySDS.
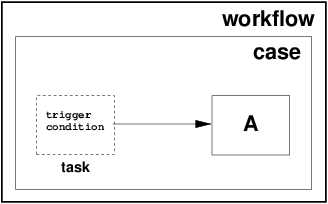
Implicit Workflow
Implicit workflow capability is a first-class, built-in feature of the HySDS architecture. HySDS provides users with access to 2 workflow data patterns that compose its implicit workflow capability:
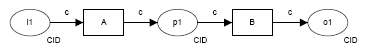
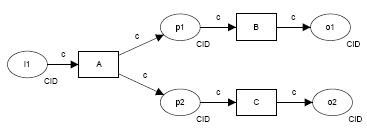
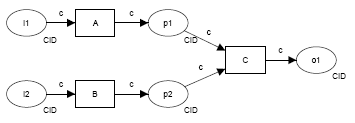
| pattern | description | diagram (courtesy http://www.workflowpatterns.com) |
|---|---|---|
| Event-based task trigger | external event initiates execution of an individual step in a workflow, e.g. S3 event triggers an AWS Lambda call via SNS to submit an ingest job | |
| Data-based task trigger |
|
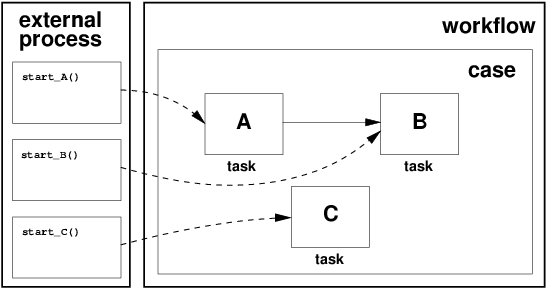
In the previous "Job Execution" section, the sequence diagram describes the Worker submitting a evaluator task to the Queue before exiting its job execution (step 14). Execution of this evaluation task by the Helper determines if there are subsequent steps (jobs) that can be initiated. If so, those jobs are submitted to the Orchestrator and the workflow continues on. By definition, workflows built in this manner are implicitly defined by the chain of trigger rules that connect job types to each other by the datasets they produce. As an example, the following diagram describes the end-to-end pipeline for the GRFN (Getting Ready for NISAR) standard product (GUNW - geocoded unwrapped interferogram):
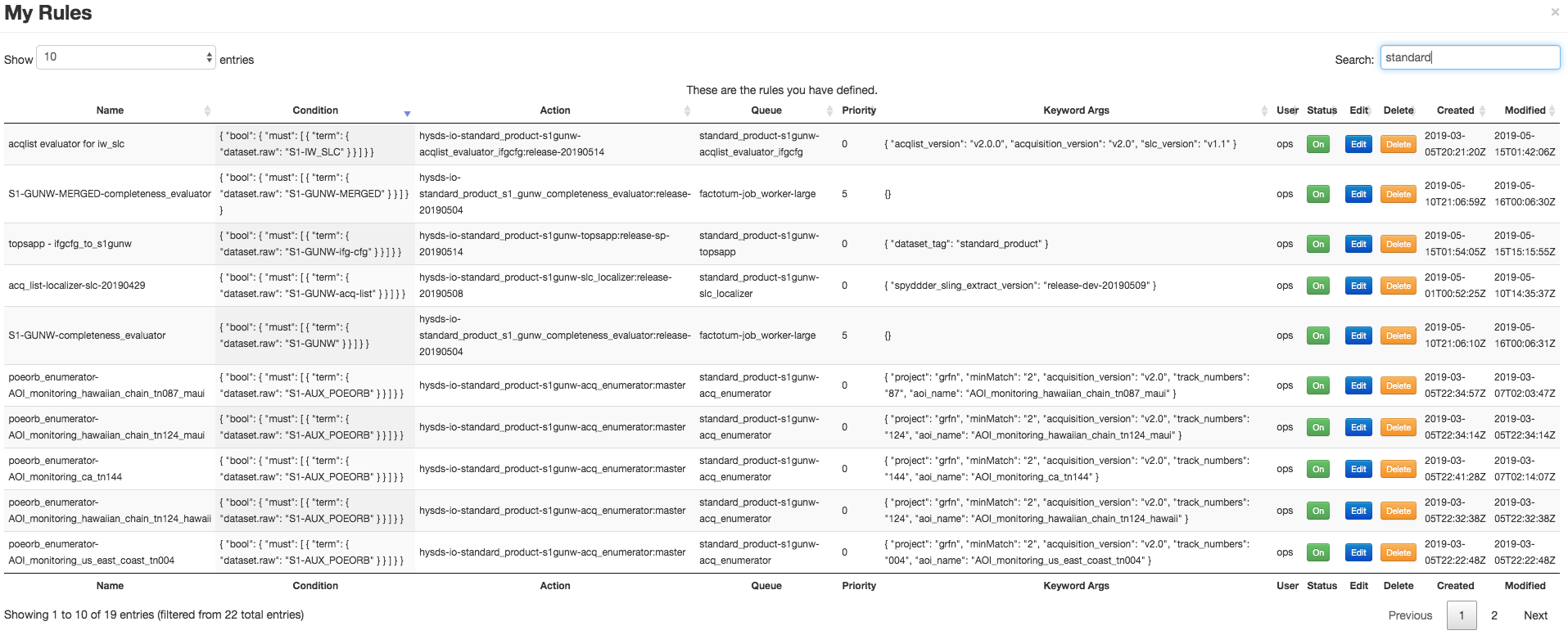
The following screenshot shows the set of trigger rules that compose the above pipeline into an implicit workflow:
Explicit Workflow
In contrast, explicit workflow capability is not a first-class, built-in feature in the HySDS architecture. Any explicit workflow functionality must be encapsulated into the base atomic unit of execution in HySDS: the job. Therefore, explicit workflow execution must be codified into a HySDS job type but this gives users the freedom to implement explicit workflows using the workflow engine of their choice. By default, HySDS provides users with built-in explicit workflow capability using the SciFlo workflow engine which provides hooks into the HySDS job submission/execution framework. Adaptation of other workflow frameworks to hook into the HySDS framework is possible using the Mozart REST API.
SciFlo subscribes to the control-flow pattern in which execution order of individual steps in a workflow are explicitly defined. SciFlo supports the following control-flow patterns:
| pattern | diagram (courtesy http://www.workflowpatterns.com) |
|---|---|
The following example shows a SciFlo workflow definition that implements a Map-Reduce job that is composed of the parallel split and synchronization patterns as described above. Map jobs submitted by the workflow to HySDS are executed in parallel and the workflow implements the blocking at the Reduce step:
<?xml version="1.0"?>
<sf:sciflo xmlns:sf="http://sciflo.jpl.nasa.gov/2006v1/sf"
xmlns:xs="http://www.w3.org/2001/XMLSchema"
xmlns:py="http://sciflo.jpl.nasa.gov/2006v1/py">
<sf:flow id="test_par_python_all">
<sf:description>Test par python for both map and reduce steps.</sf:description>
<sf:inputs>
<startdate type="xs:date">2010-01-01T00:00:00Z</startdate>
<enddate type="xs:date">2010-12-31T23:59:59Z</enddate>
<segmentBy type="xs:int">31</segmentBy>
<arg1 type="xs:string">Hello</arg1>
<arg2 type="xs:string">World</arg2>
</sf:inputs>
<sf:outputs>
<res1>@#get_reduce_result</res1>
<res2>@#get_reduce_result_async</res2>
</sf:outputs>
<sf:processes>
<sf:process id="segment_months">
<sf:inputs>
<startdate/>
<enddate/>
<segmentBy/>
</sf:inputs>
<sf:outputs>
<seg_startdate/>
<seg_enddate/>
<seg_days/>
</sf:outputs>
<sf:operator>
<sf:description></sf:description>
<sf:op>
<sf:binding>python:?sciflo.mapreduce.test.date_segmenter</sf:binding>
</sf:op>
</sf:operator>
</sf:process>
<sf:process id="map">
<sf:inputs>
<seg_startdate from="@#previous" />
<seg_enddate from="@#previous" />
<seg_days from="@#previous" />
<arg1/>
<arg2/>
</sf:inputs>
<sf:outputs>
<mapped_results/>
</sf:outputs>
<sf:operator>
<sf:description></sf:description>
<sf:op>
<sf:binding job_queue="jobs_processed">map:python:?sciflo.mapreduce.test.create_map_job</sf:binding>
</sf:op>
</sf:operator>
</sf:process>
<sf:process id="reduce">
<sf:inputs>
<mapped_results from="@#previous" />
</sf:inputs>
<sf:outputs>
<reduced_result/>
</sf:outputs>
<sf:operator>
<sf:description></sf:description>
<sf:op>
<sf:binding job_queue="jobs_processed">parallel:python:?sciflo.mapreduce.test.create_reduce_job</sf:binding>
</sf:op>
</sf:operator>
</sf:process>
<sf:process id="get_reduce_result">
<sf:inputs>
<reduced_result from="@#previous" />
</sf:inputs>
<sf:outputs>
<res/>
</sf:outputs>
<sf:operator>
<sf:description></sf:description>
<sf:op>
<sf:binding>python:?sciflo.mapreduce.test.get_reduce_job_result</sf:binding>
</sf:op>
</sf:operator>
</sf:process>
<sf:process id="map_async">
<sf:inputs>
<seg_startdate from="@#segment_months" />
<seg_enddate from="@#segment_months" />
<seg_days from="@#segment_months" />
<arg1/>
<arg2/>
</sf:inputs>
<sf:outputs>
<mapped_results/>
</sf:outputs>
<sf:operator>
<sf:description></sf:description>
<sf:op>
<sf:binding job_queue="jobs_processed" async="true">map:python:?sciflo.mapreduce.test.create_map_job</sf:binding>
</sf:op>
</sf:operator>
</sf:process>
<sf:process id="reduce_async">
<sf:inputs>
<mapped_results from="@#previous" />
</sf:inputs>
<sf:outputs>
<reduced_results/>
</sf:outputs>
<sf:operator>
<sf:description></sf:description>
<sf:op>
<sf:binding>python:?sciflo.mapreduce.test.join_map_jobs</sf:binding>
</sf:op>
</sf:operator>
</sf:process>
<sf:process id="get_reduce_result_async">
<sf:inputs>
<reduced_result from="@#previous" />
</sf:inputs>
<sf:outputs>
<res/>
</sf:outputs>
<sf:operator>
<sf:description></sf:description>
<sf:op>
<sf:binding>python:?sciflo.mapreduce.test.reduce_map_jobs</sf:binding>
</sf:op>
</sf:operator>
</sf:process>
</sf:processes>
</sf:flow>
</sf:sciflo>
Consult the SciFlo github repository at https://github.com/hysds/sciflo for more detailed information.
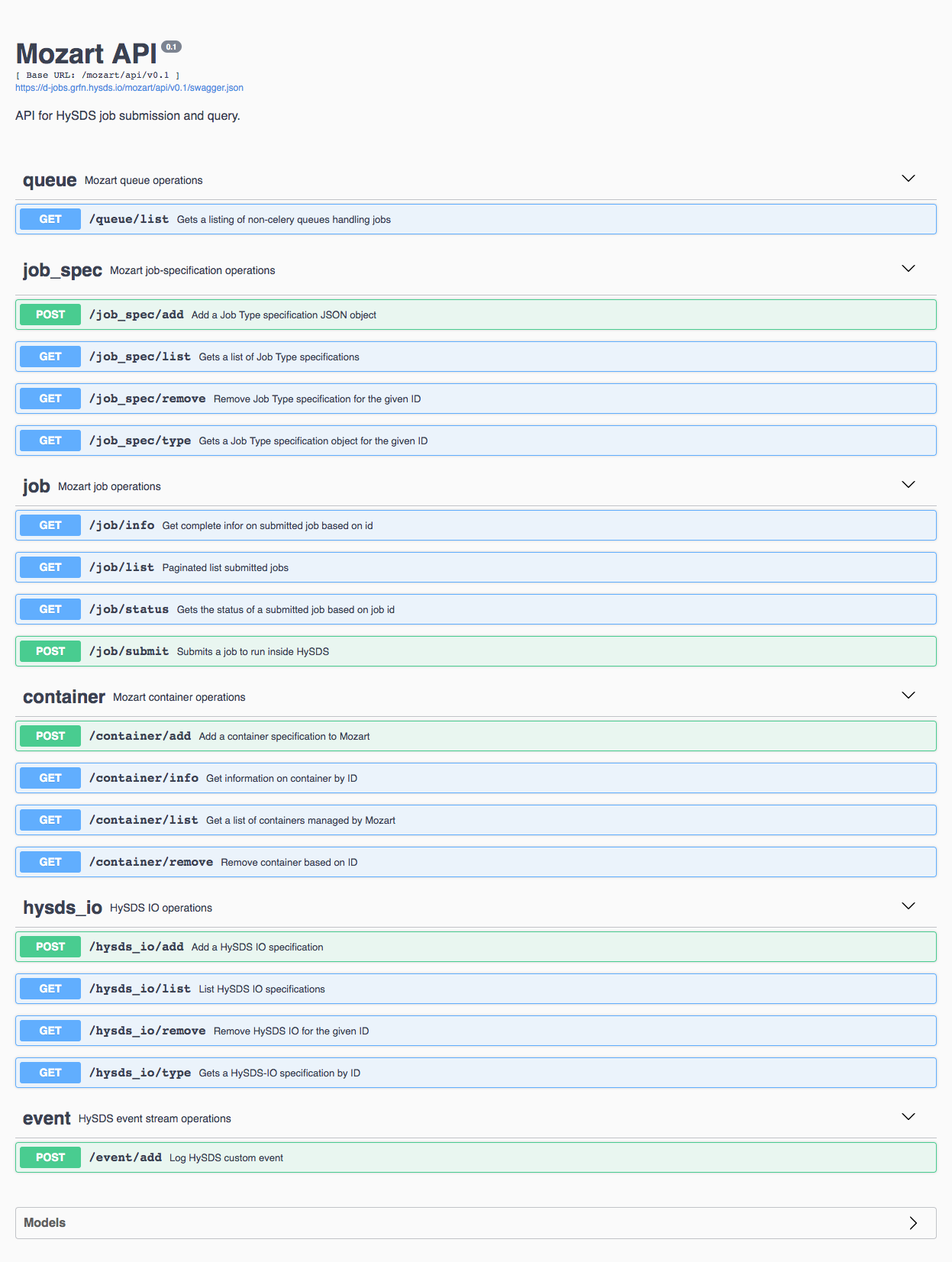
Resource Management REST API (mozart)
HySDS provides a REST application programming interface (API) as part of the resource management subsystem. This API conforms to the OpenAPI specification (formerly Swagger) and provides REST interfaces for performing operations on queues, job-specs, jobs, containers, hysds-ios and events.
Queue Operations
| uri | method | description |
|---|---|---|
| /queue/list | GET | Get a listing of non-celery queues handling jobs |
job-spec Operations
| uri | method | description |
|---|---|---|
| /job_spec/add | POST | Add a job type specification JSON object |
| /job_spec/list | GET | Get a list of job type specifications |
| /job_spec/remove | GET | Remove job type specification for the given ID |
| /job_spec/type | GET | Get a job type specification object for the given ID |
job Operations
| uri | method | description |
|---|---|---|
| /job/info | GET | Get complete info on submitted job based on ID |
| /job/list | GET | Paginated list of submitted jobs |
| /job/status | GET | Get the status of a submitted job based on job ID |
| /job/submit | POST | Submit a job to run on HySDS |
container Operations
| uri | method | description |
|---|---|---|
| /container/add | POST | Add a container specification |
| /container/info | GET | Get information on container by ID |
| /container/list | GET | Get a list of containers |
| /container/remove | GET | Remove container based on ID |
hysds-io Operations
| uri | method | description |
|---|---|---|
| /hysds_io/add | POST | Add a HySDS IO specification |
| /hysds_io/list | GET | List HySDS IO specifications |
| /hysds_io/remove | GET | Remove HySDS IO for the given ID |
| /hysds_io/type | GET | Get a HySDS IO specification by ID |
event Operations
| uri | method | description |
|---|---|---|
| /event/add | POST | Log HySDS custom event |
Interface Description:
Communication media and protocols - REST over HTTP/HTTPS protocol
Format - Input is performed via either HTTP GET or POST actions. Inputs are in the for of URL addressing with query parameters for GET actions. For POST actions, input parameters are encoded in the HTTP request body. The results, if any, are returned as JSON formatted objects.
How to Detect and Handle Errors - Errors in the REST call are indicated by the returned HTTP response code.
The following screenshot shows the browser-based UI provided by the Mozart REST API. This UI provides both documentation and an interactive interface to call the REST endpoints.
Resource Management UI (figaro)
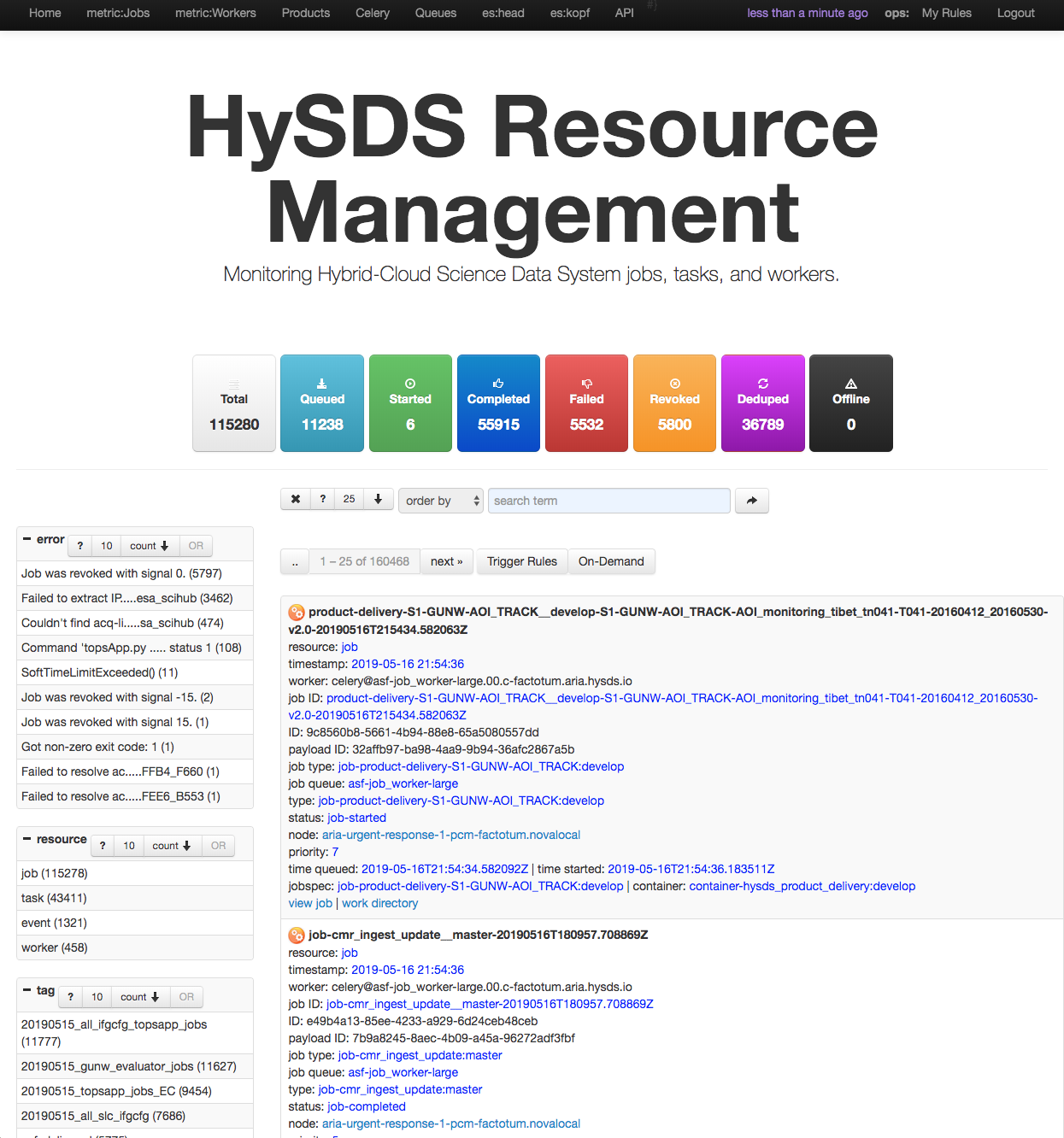
HySDS provides a web user interface for the user to perform the following functions:
- Manual management of job execution (submit, revoke, purge)
- Monitoring job execution
- Monitoring task execution
- Monitoring work execution
- Monitoring events
This resource management UI, also known as figaro, is a custom built web application adapted from the FacetView project. It provides a faceted search interface allowing users to narrow down search results based on the application of multiple filters.
Interface Description:
Communication media and protocols - Browser-based interactive web app. The communication protocol is over secured Hypertext Transfer Protocol (HTTPS).
Format - For job management, input is text/string based and is entered via various web forms. For searching, input is a combination of typing text/string into a search input field and clicking on facet values.
How to Detect and Handle Errors - The web app will display interface, data entry, and internal server errors on the application web page. The user decides how to best handle any resulting errors.
Screenshot of the Resource Management UI is shown below:
Dataset Management REST API (grq2)
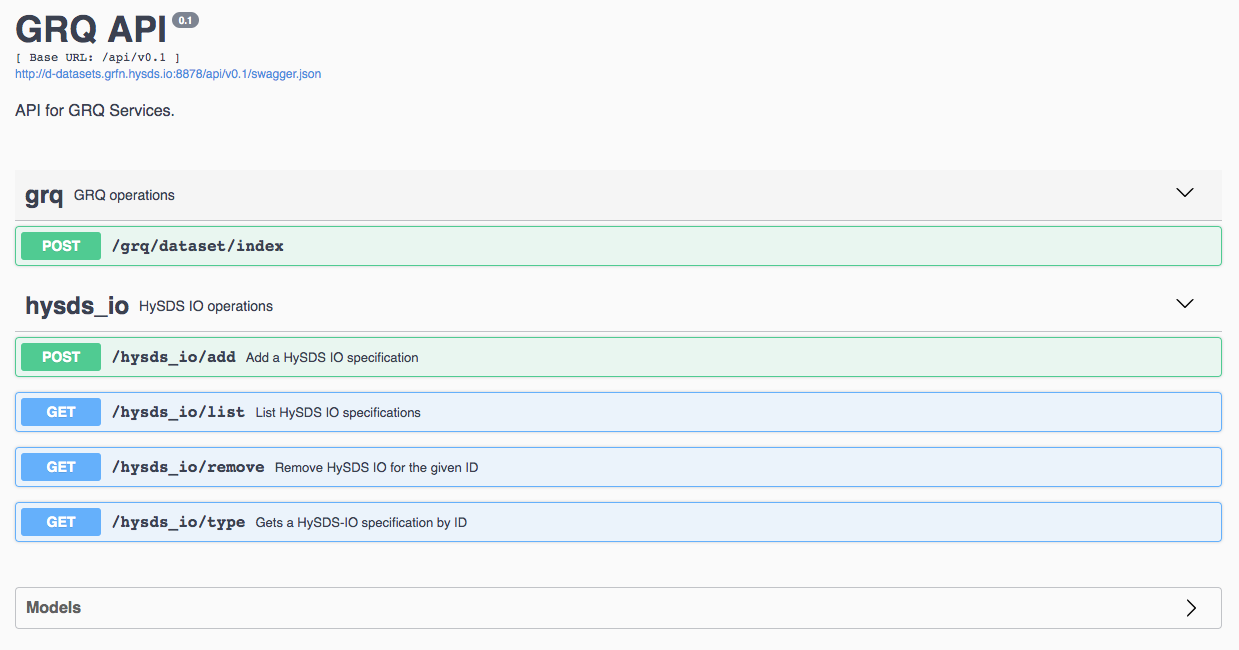
HySDS provides a REST application programming interface (API) as part of the Discovery subsystem. This API conforms to the OpenAPI specification (formerly Swagger) and provides REST interfaces for performing operations on hysds-ios installed in the Discovery subsystem as well as indexing datatasets into the catalog.
dataset Operations
| uri | method | description |
|---|---|---|
| /grq/dataset/index | POST | Index dataset |
hysds-io Operations
| uri | method | description |
|---|---|---|
| /hysds_io/add | POST | Add a HySDS IO specification |
| /hysds_io/list | GET | List HySDS IO specifications |
| /hysds_io/remove | GET | Remove HySDS IO for the given ID |
| /hysds_io/type | GET | Get a HySDS IO specification by ID |
Interface Description:
Communication media and protocols - REST over HTTP/HTTPS protocol
Format - Input is performed via either HTTP GET or POST actions. Inputs are in the for of URL addressing with query parameters for GET actions. For POST actions, input parameters are encoded in the HTTP request body. The results, if any, are returned as JSON formatted objects.
How to Detect and Handle Errors - Errors in the REST call are indicated by the returned HTTP response code.
The following screenshot shows the browser-based UI provided by the GRQ REST API. This UI provides both documentation and an interactive interface to call the REST endpoints.
Dataset Facet Search UI (tosca)
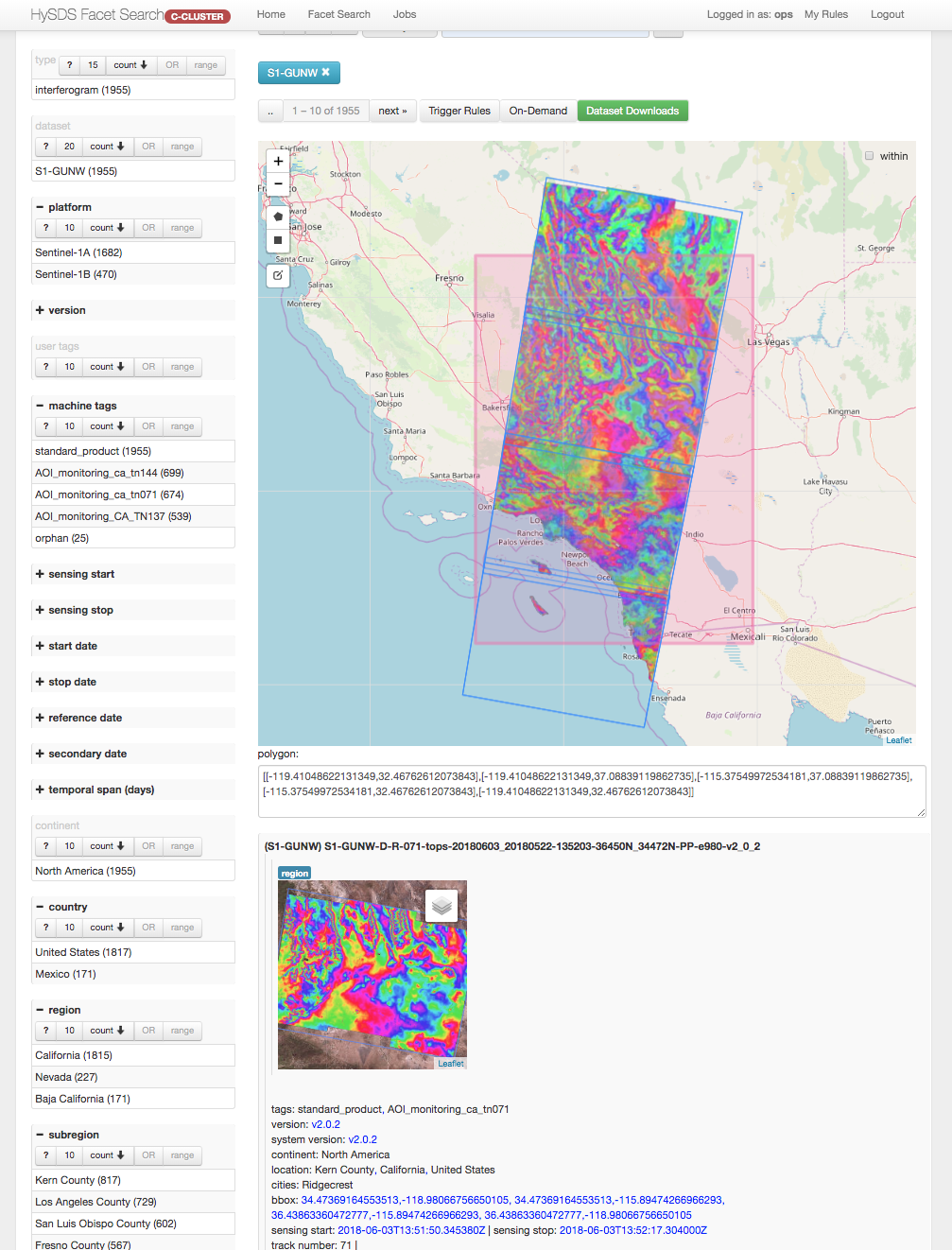
HySDS provides a web user interface for the user to perform the following functions:
- Facet search on datasets ingested and/or published into the system
- Manual management of job datasets (purge, generate wget script)
- Submit on-demand jobs
- CRUD management of trigger rules for job types
This resource management UI, also known as tosca, is a custom built web application adapted from the FacetView project. It provides a faceted search interface allowing users to narrow down search results based on the application of multiple filters.
Interface Description:
Communication media and protocols - Browser-based interactive web app. The communication protocol is over secured Hypertext Transfer Protocol (HTTPS).
Format - input is text/string based and is entered via various web forms. For searching, input is a combination of typing text/string into a search input field and clicking on facet values.
How to Detect and Handle Errors - The web app will display interface, data entry, and internal server errors on the application web page. The user decides how to best handle any resulting errors.
Screenshot of the Dataset Facet Search UI is shown below:
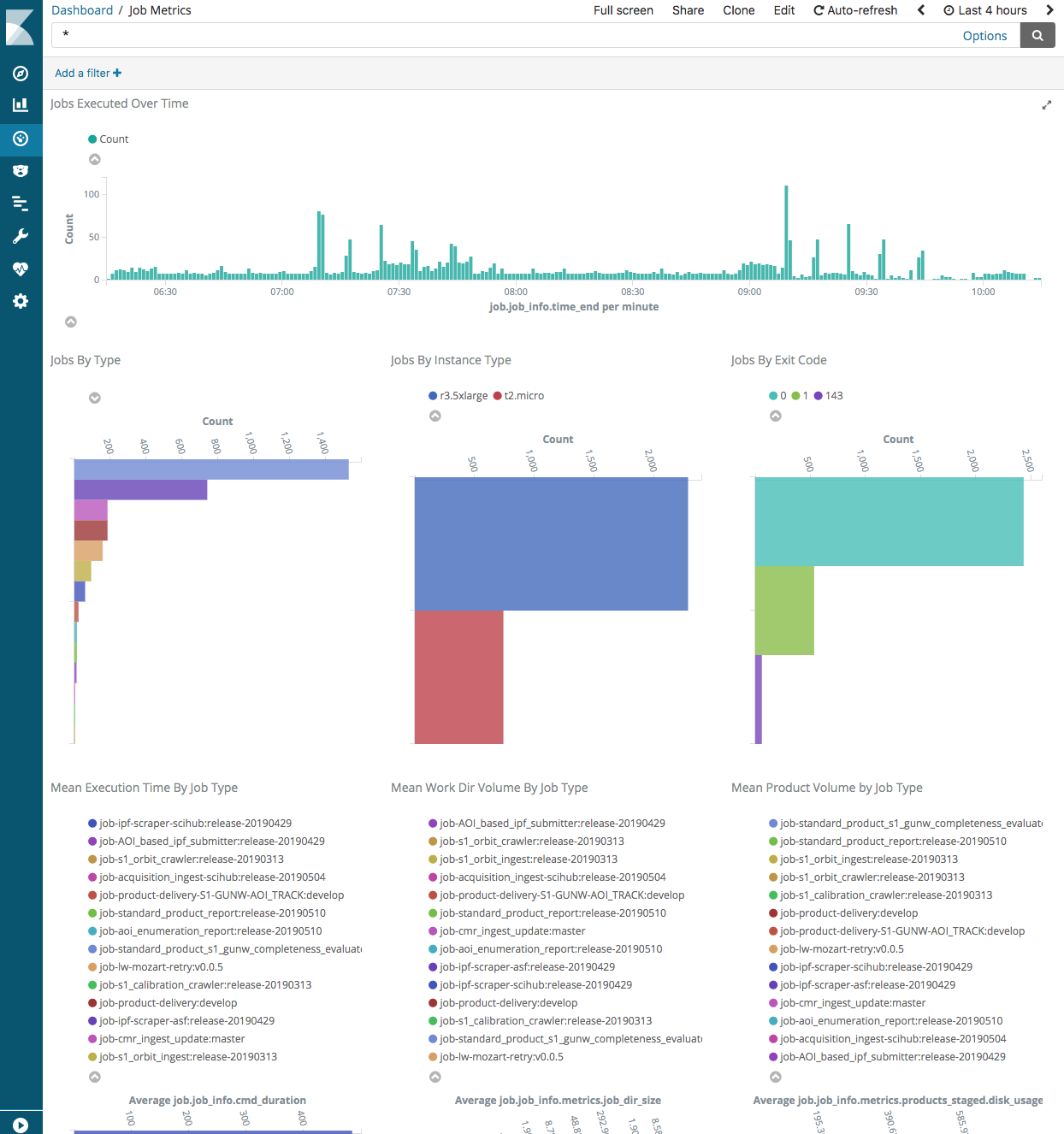
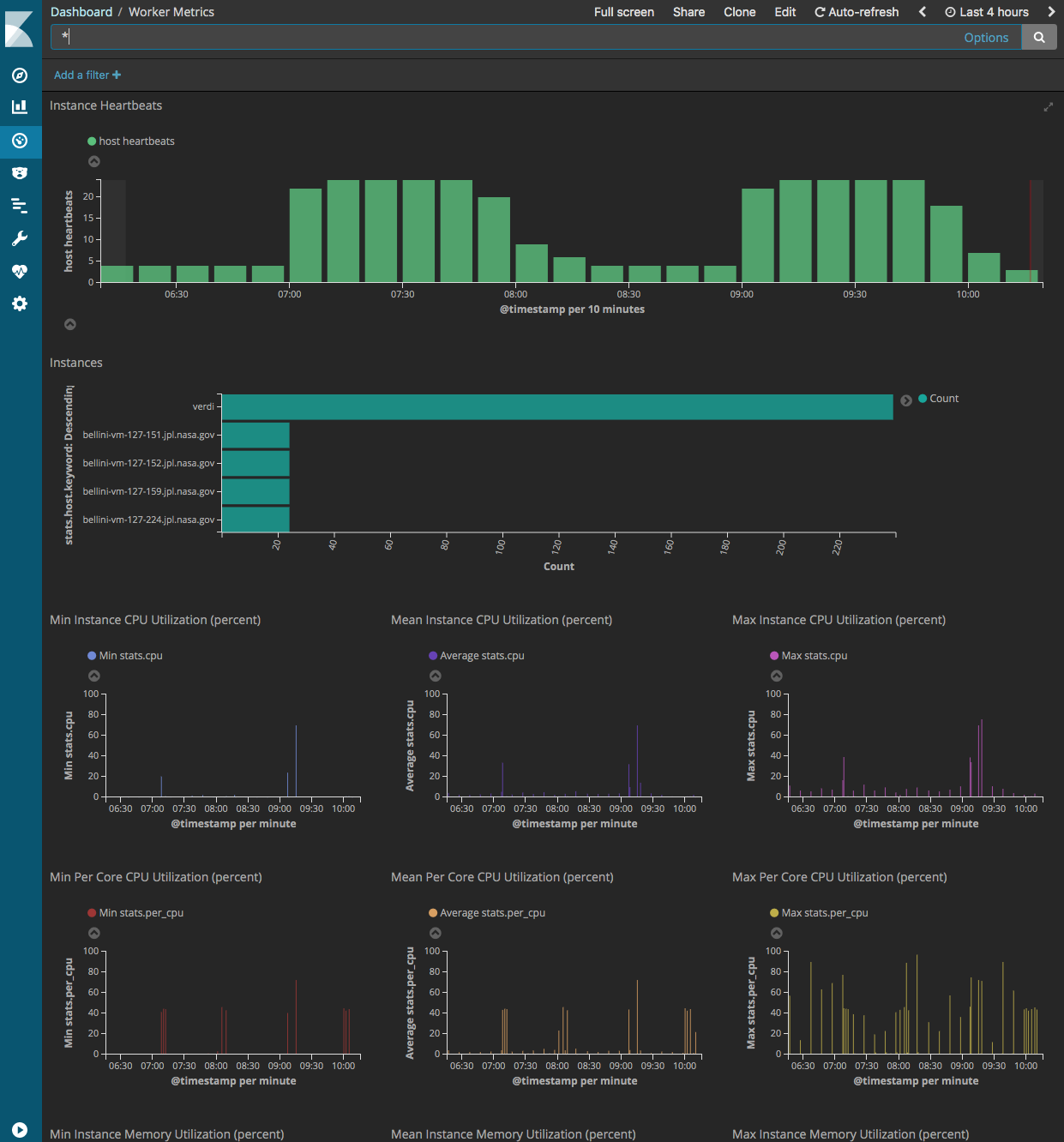
Job and Worker Metrics Dashboard (kibana)
HySDS provides a web user interface for the user to perform the following functions:
- Monitor job metrics
- Monitor worker metrics
These metrics dashboards are built using the Kibana visualization web application. It provides a faceted search interface allowing users to narrow down search results based on the application of multiple filters.
Interface Description:
Communication media and protocols - Browser-based interactive web app. The communication protocol is over secured Hypertext Transfer Protocol (HTTPS).
Format - input is text/string based and is entered via various web forms. For searching, input is a combination of typing text/string into a search input field and clicking on facet values.
How to Detect and Handle Errors - The web app will display interface, data entry, and internal server errors on the application web page. The user decides how to best handle any resulting errors.
Screenshots of the job and worker metrics dashboards are shown below:
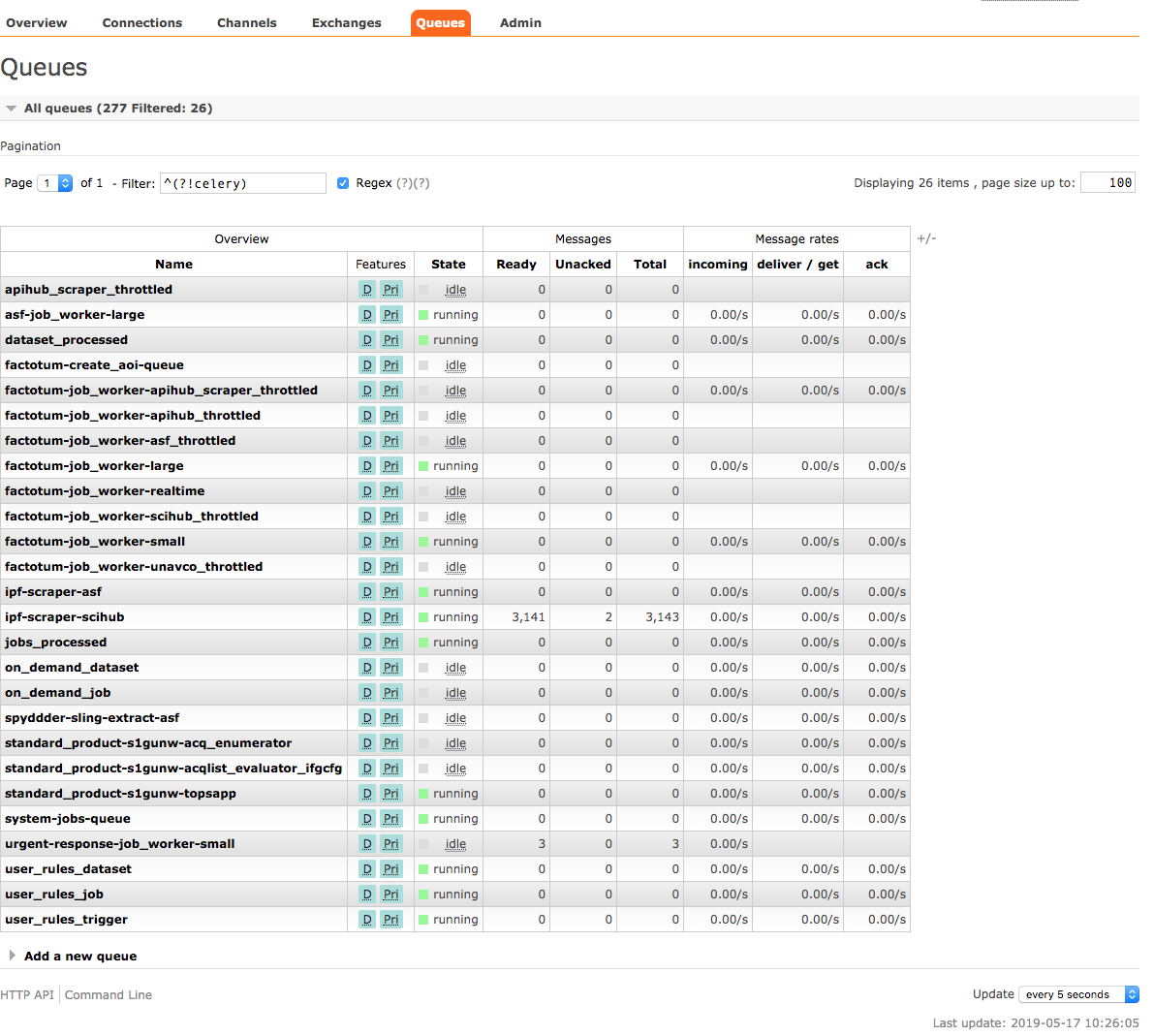
RabbitMQ Admin Interface
HySDS provides a web user interface for the user to perform the following functions:
- Manual management of RabbitMQ exchanges, queues, channels and connections
- Monitoring queues and the status of messages on the queues
This RabbitMQ administration interface is provided out-the-box by the RabbitMQ admin plugin.
Interface Description:
Communication media and protocols - Browser-based interactive web app. The communication protocol is over secured Hypertext Transfer Protocol (HTTPS).
Format - input is text/string based and is entered via various web forms. For searching, input is a combination of typing text/string into a search input field.
How to Detect and Handle Errors - The web app will display interface, data entry, and internal server errors on the application web page. The user decides how to best handle any resulting errors.
Screenshot of the RabbitMQ admin interface is shown below:
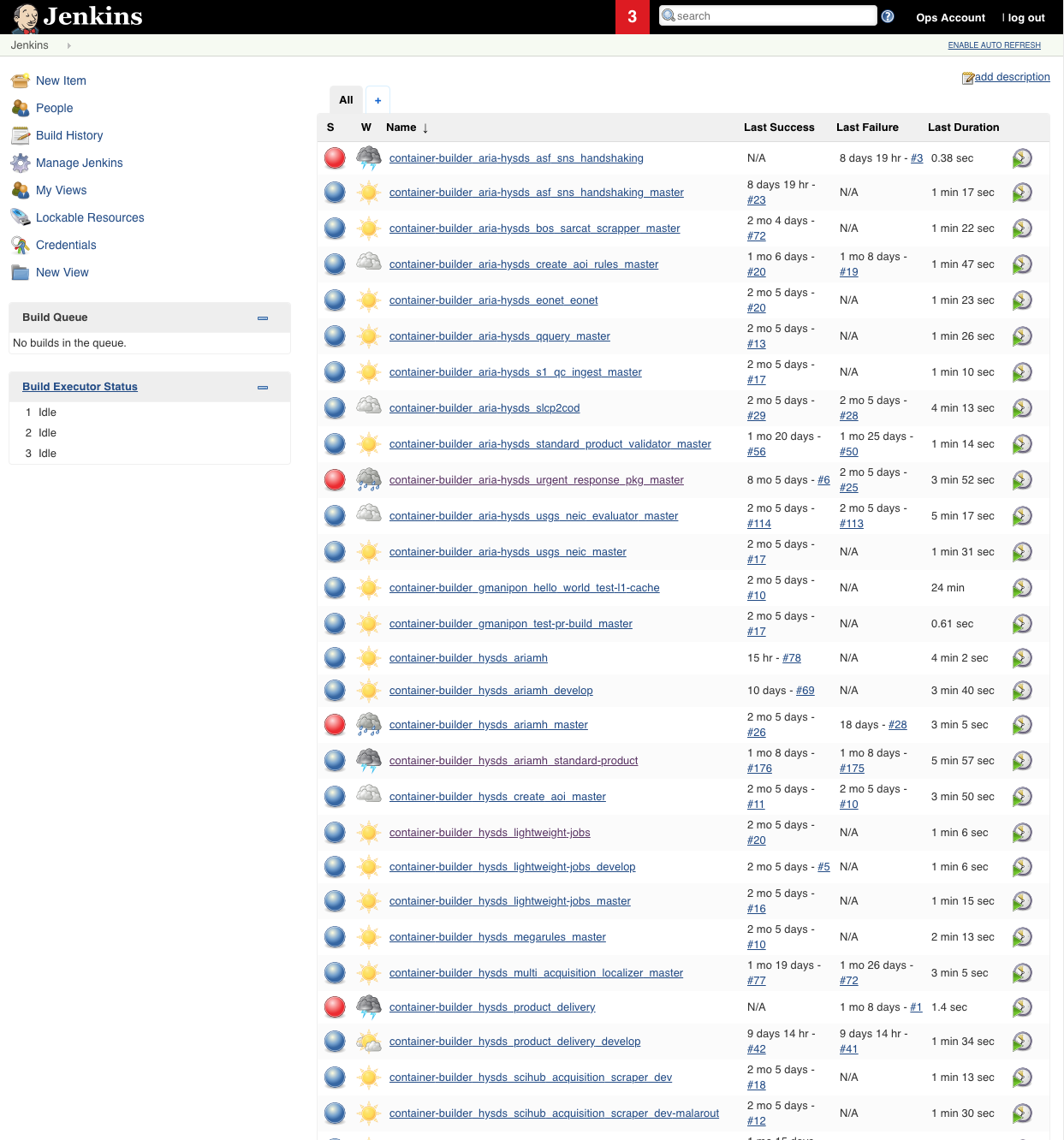
Jenkins
HySDS provides a web user interface for the user to perform the following functions:
- Manual management of HySDS job container builds
- Monitoring of job container build execution and progress
This interface is provided out-the-box by the Jenkins web application.
Interface Description:
Communication media and protocols - Browser-based interactive web app. The communication protocol is over secured Hypertext Transfer Protocol (HTTPS).
Format - input is text/string based and is entered via various web forms. For searching, input is a combination of typing text/string into a search input field.
How to Detect and Handle Errors - The web app will display interface, data entry, and internal server errors on the application web page. The user decides how to best handle any resulting errors.
Screenshot of the Jenkins interface is shown below:
HySDS Services
This section describes various services and backend components that HySDS implements. These services are needed for HySDS to function.
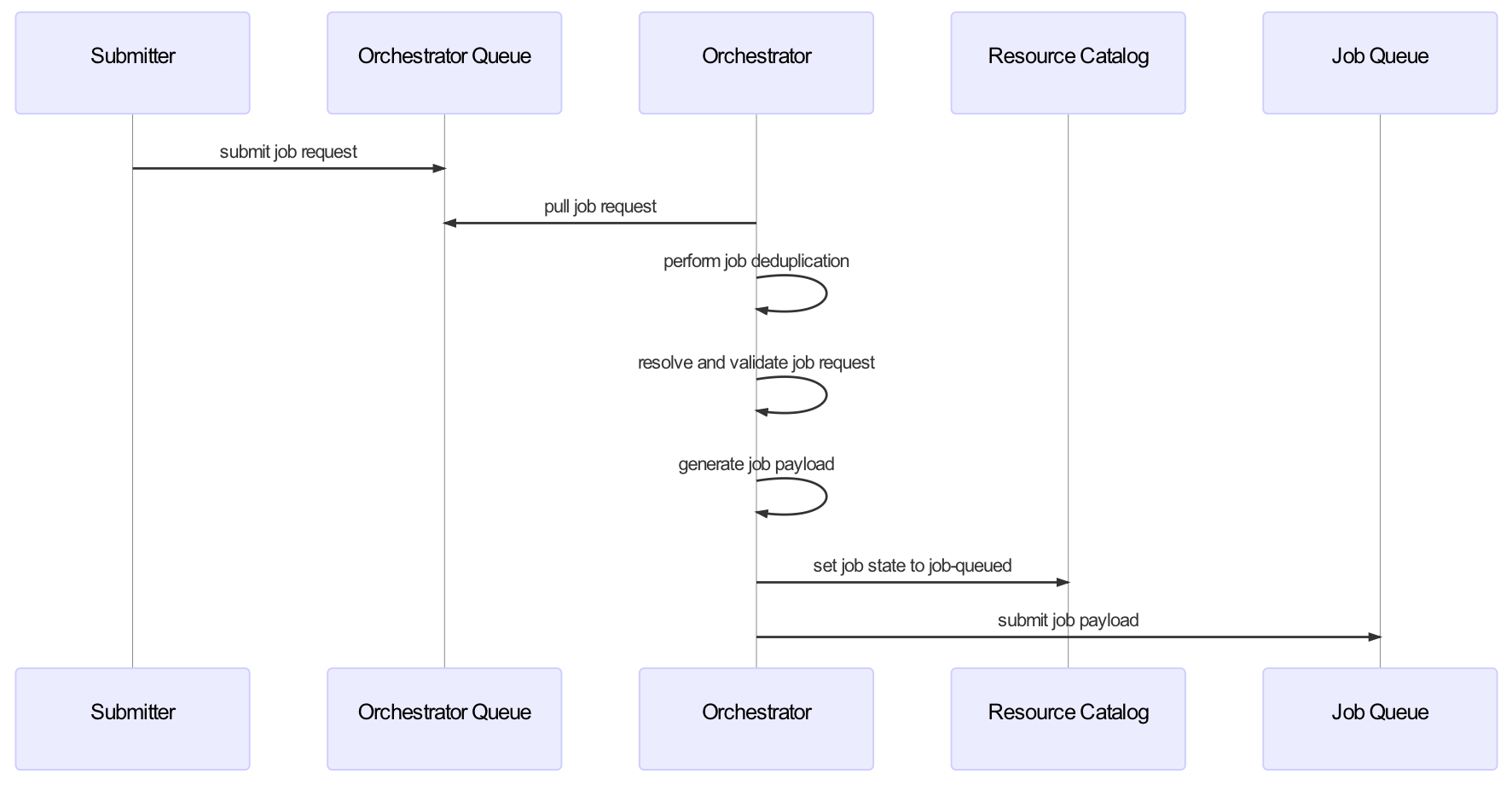
Orchestrator
The orchestrator, as depicted in the diagram below, plays a central role in the submission of jobs into the HySDS cluster. It serves as a gateway to validate and register job submission requests before handing them off to the workers (verdi) that will run the actual jobs.
The orchestrator is itself a celery worker and the following sequence diagram describes the series of events that it performs.
Helper Worker (factotum)
As stated previously, the helper subsystem (factotum) is a specialization of the compute service subsystem (verdi) that encapsulates the various 3rd party software and HySDS components related to the execution of system-based HySDS jobs and lightweight Celery tasks.
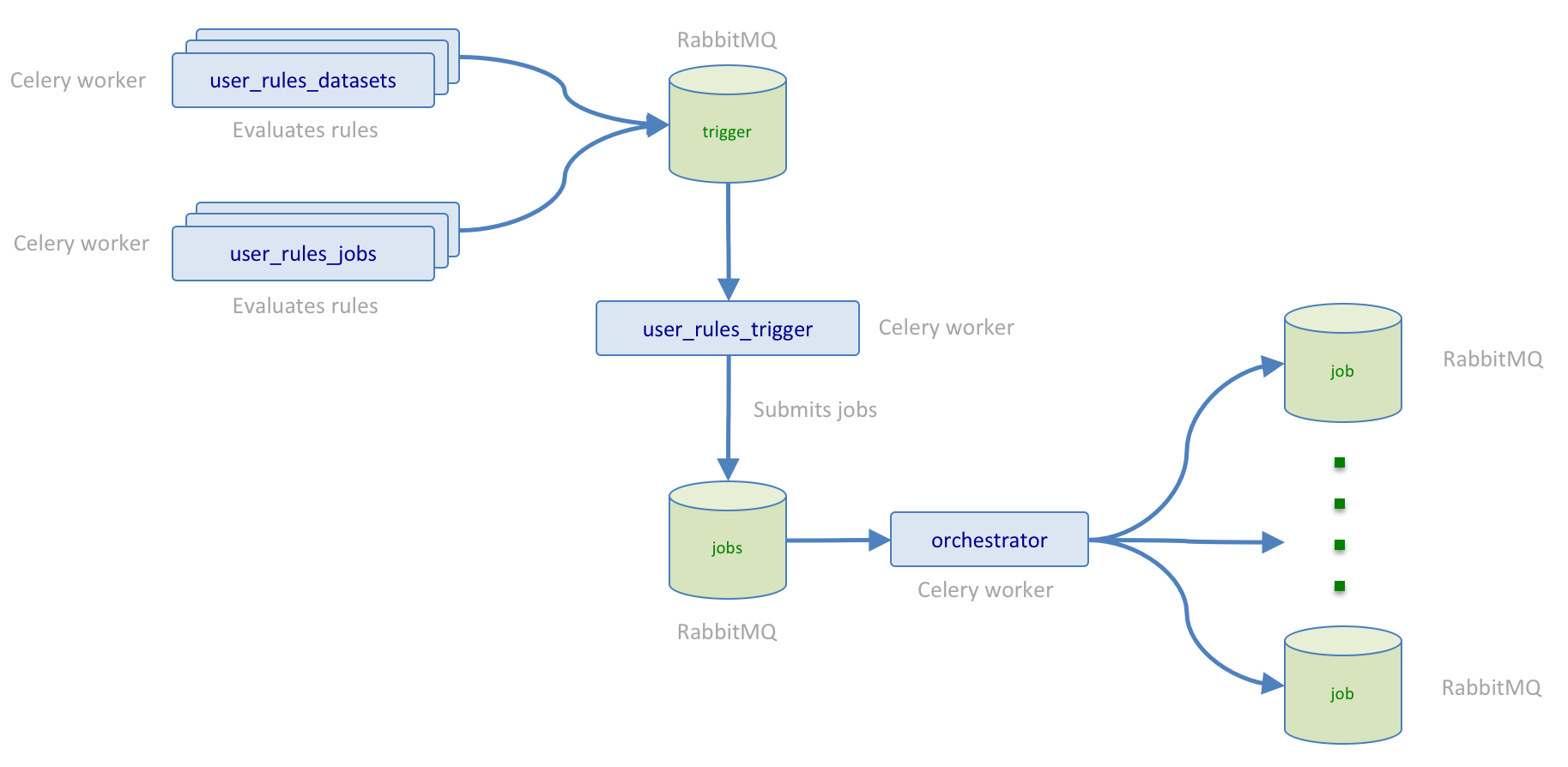
The helper subsystem plays a central role in the execution of jobs and tasks related to trigger rule evaluation. The diagram below depicts user_rules_datasets and user_rules_jobs celery workers running in the helper subsystem that perform the evaluation of trigger rules. Whenever trigger rule evaluation results in a match, the user_rules_dataset/user_rules_jobs worker submits a task for the user_rules_trigger celery workers also running in the helper subsystem to submit a job request to the orchestrator.
Analysis
Imported Software
As previously discussed, the following diagram identifies the major technologies and software components utilized by HySDS. See the implementation diagram in a previous section to view the dependency tree of HySDS core components to 3rd party software.
Design Feasibility, Performance and Margins
The HySDS design has been shown to be feasible based on initial prototypes for the ARIA, GRFN and WVCC projects. In addition, the GRFN project's HySDS cluster was able to scale up to 8000 worker (verdi) nodes exercising the scalability of the system.
Based on these initial prototyping and testing efforts, it is determined that the performance of the HySDS design is adequate for a majority of the HySDS uses. Although more effort is required to define formal performance requirements, HySDS is purposefully designed to provide a scalable system for those use cases that require a high throughput and low latency system.
Reliability, Maintainability, and Related Requirements
Reliability of HySDS is important, as it will be used to support applications that require a high level of reliability. The reliability aspects of HySDS have been considered in the selection and use of Celery & RabbitMQ as the base distributed task framework and message broker, respectively. Part of the effort in upcoming development cycles is to formally define these reliability requirements. The reliability of HySDS will be tracked, and the design will be updated if reliability needs are not met.
Maintainability of HySDS is at the forefront of the design. HySDS has a self- imposed constraint that prevents the forking of open-source third party software libraries to implement HySDS functionality. This is done to promote the maintainability of the code. Additionally, the design of the HySDS custom developed software has gone through several iterations such that they are architected in a loosely coupled way to promote a highly cohesive system.
Adaptability and Reusability
The HySDS design provides several points of configurability and extension points, which allows it to be adaptable for specific mission requirements. The goal of HySDS is to provide a scalable and reusable science data system. The re-usable core software is managed as an open source project and is available to the public for use.
SWOT-specific Adaptation
Overview
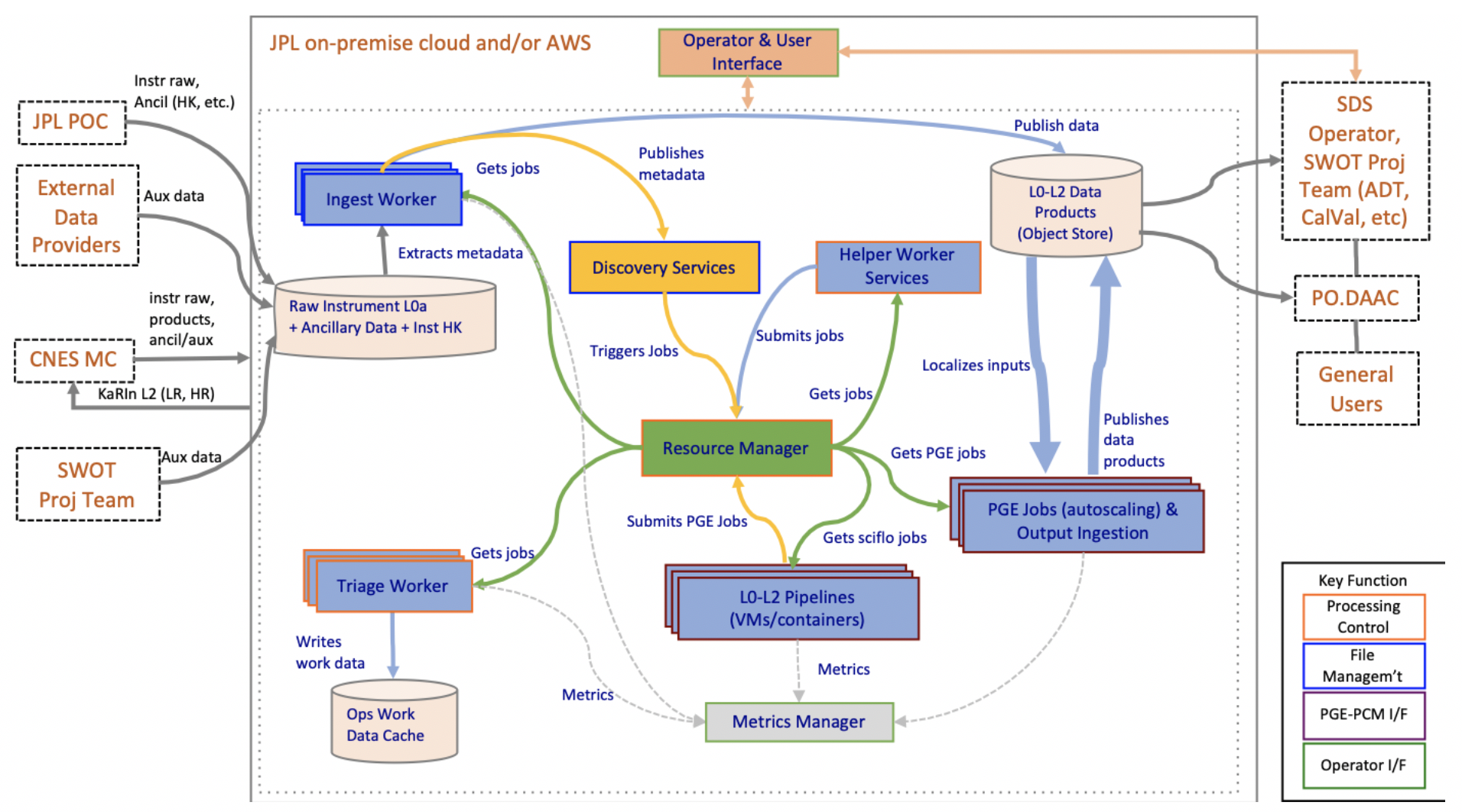
The SWOT Processing Control & Management (PCM) subsystem extends the functionality provided by HySDS and contains the following functions:
- Ingest, catalog, and store mission data and ancillary datasets enabling science data processing
- Ingest mission products retrieved from PO.DAAC, in support of bulk reprocessing
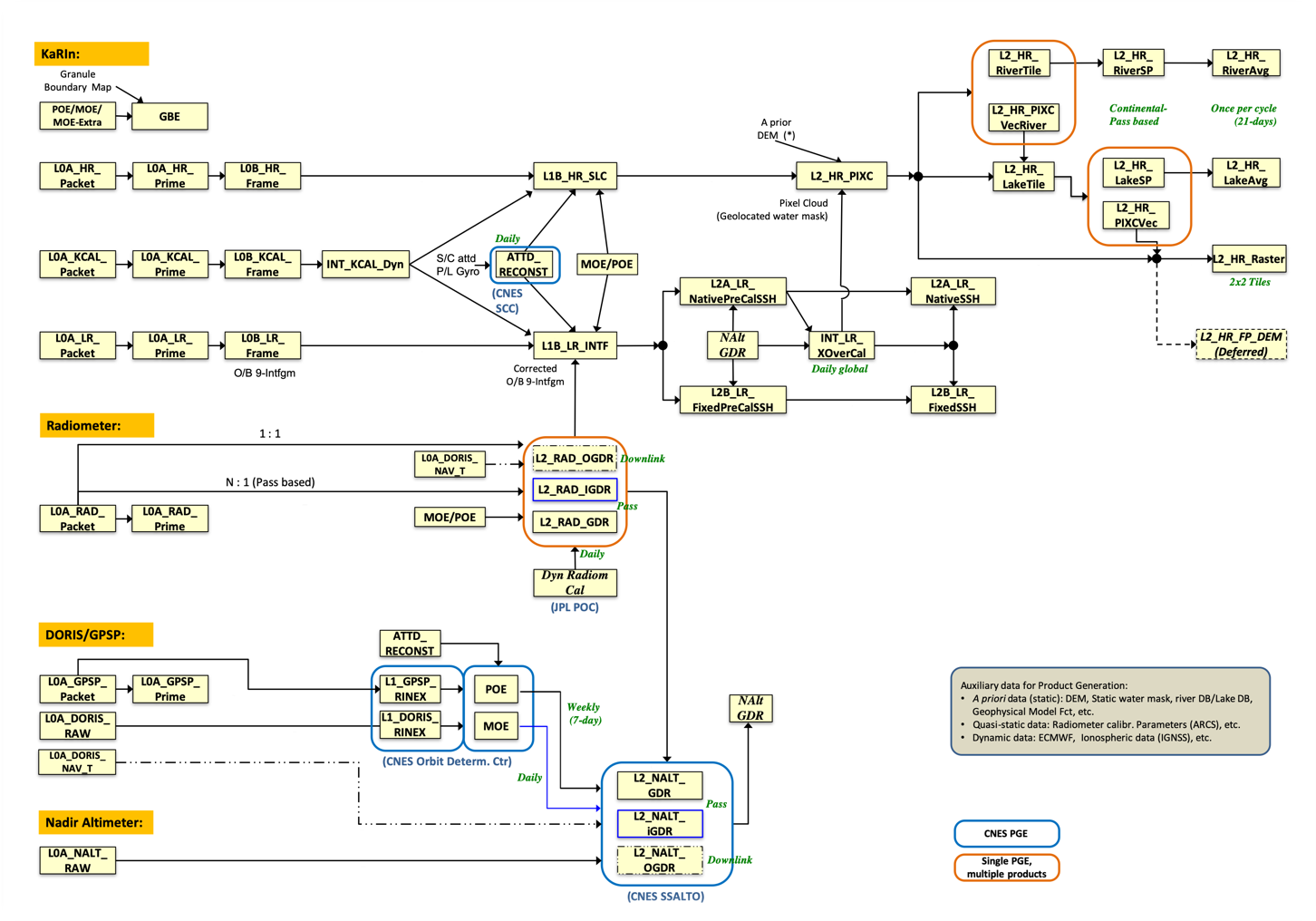
- Automate science data processing to produce Level 0 through Level 2 KaRIn, AMR (Radiometer), GPS data products, scaling the PCM’s distributed processing system as needed
- Interface with L0-L2 PGEs and Ancillary Preprocessors
- Store and catalog L0-L2 science data products and their associated files
- Monitor data system services, PGE executions, and computing resources by providing operators with a user interface for the PCM system
- Report data processing and product accounting
- Make available the science data products for DAAC within the latency requirements
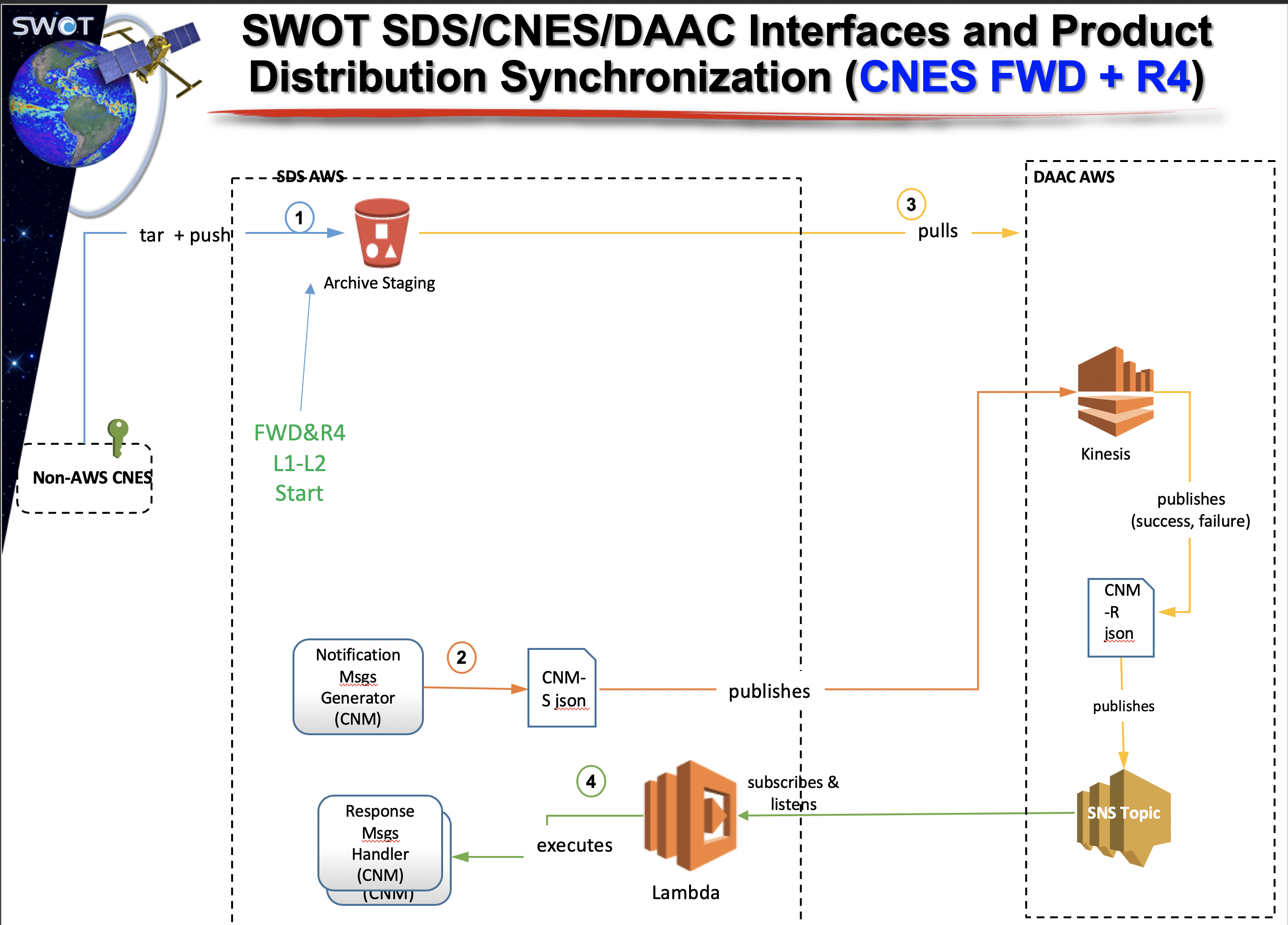
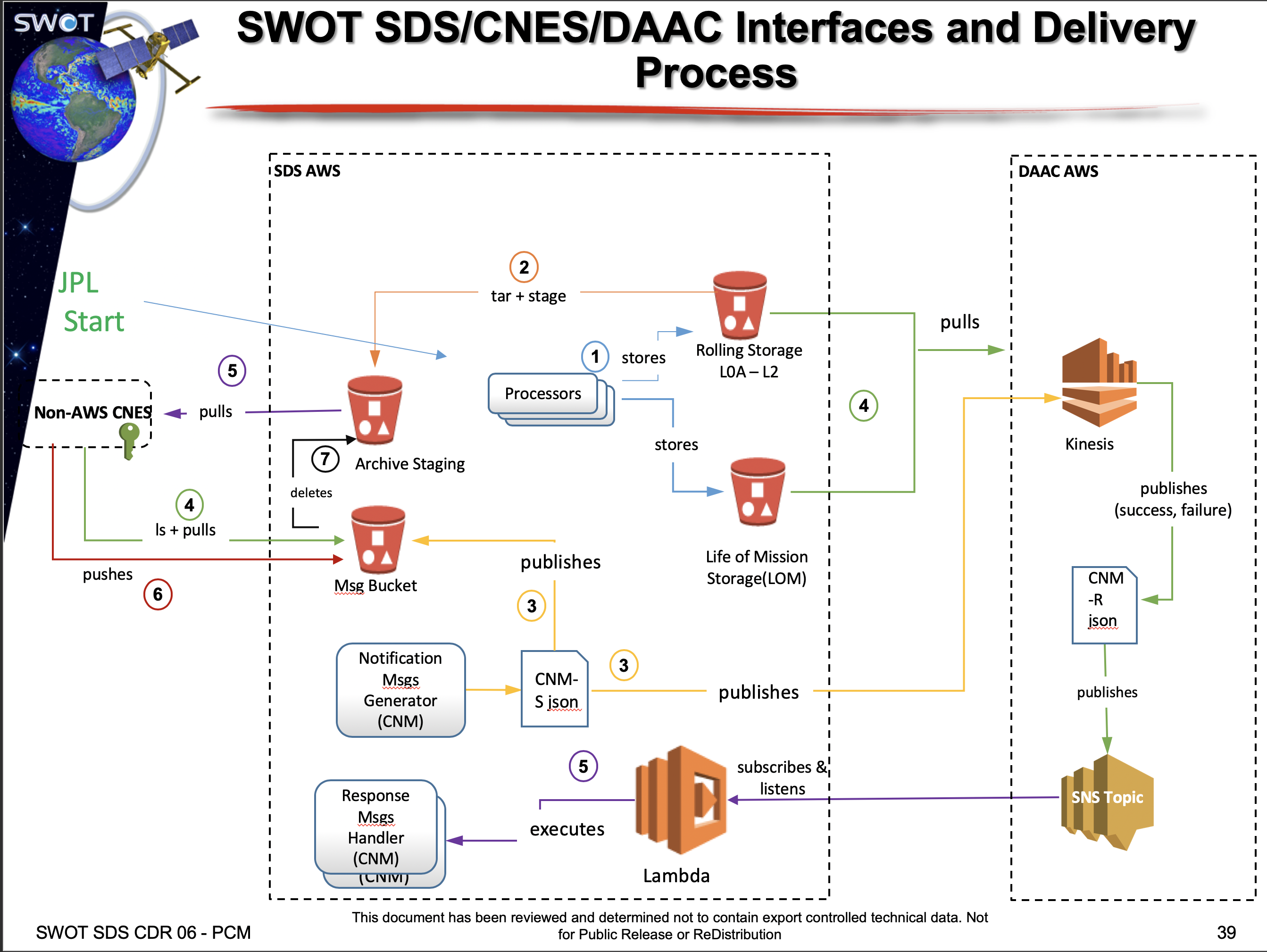
- Make available data products for CNES within the latency requirements
- Provide data access to the Science Team
To fulfill the operations need, the PCM is designed to operates automatically 24 hours a day and seven days a week.
The major components of PCM include the following:
- Resource Manager
- Workflow Manager
- Worker Manager
- Discovery Services
- Metrics Manager
- Helper Worker Services
- Operator Interfaces
Through the adaptation of PCM core function based on HySDS framework, PCM plugs in the SWOT specific functions including the following:
- PGE-PCM Interfaces
- External Interfaces to CNES, GDS, and PO.DAAC
- Customized Operator Interfaces
- Timer support for data gap processing
- GBE catalog
- Product to dataset conversion
- SWOT-specific reports & metrics collection
The following functional diagram illustrates the PCM components as listed above:
The HySDS adaptation of SWOT PCM includes the integration SWOT PGEs into the following processing flow:
The Resource Manager (RM)
PCM provides resource management function to SDS for computing power, storage, and their status. This component provisions, manages, and orchestrates computing instances, manages jobs, hosts user interfaces for viewing job status. The RM stores job status information, persists job database beyond PCM services shutdown.
The auto-scaling of computing resources are based on AWS’s Auto Scale feature. This is configured to dynamically allocate and scale the computing instances to support the dynamic variation of daily processing loads.
The Workflow Manager (WM)
The Workflow Manager handles the processes required to convert incoming telemetry into the requisite science data products. The workflow manager handles the complex order of processes that must operate, as well as the preconditions that must be met to initiate each process. The SWOT SDS data flow in Figure 3‑1 displays the relationship and order of processes that the SWOT PCM employs to generate Level 0 through Level 2 data products.
The WM handles daily forward processing pipelines. This component automates to trigger workflows based on input dataset rules. The workflows are also automated with time limit to trigger to meet latency requirements. There are daily ~,500 FWD processing jobs and up to 133 K jobs/day during peak bulk reprocessing campaign with the equivalent numbers of data products being generated.The WM supports on-demand and bulk reprocessing.
The WM leveraged from HySDS is based on distributed task/job queue. The jobs are executed on the worker nodes. The WM orchestrates workers that pulls PGE jobs from the queue, executes them, and publishes products. The WM also handles when job processing is interrupted, such as due to spot termination or hardware failures. The system ensures that interrupted jobs are re-queued and processed elsewhere, triaging failed jobs after a set number of retries. This allows operators to investigate failures.
The Discovery Services (DS)
File management is PCM major function. The DS catalogs products and metadata. This component also provides the user’s accesses to view product metadata and browse products. The facet searches support the generation of reports about products and the queries based on defined catalog metadata fields (e.g. product types, granule based, temporal, spatial, etc.)
The Metrics Manager (MM)
For monitoring and reporting purposes, PCM collects and displays worker and job metrics. This component hosts metrics user interface for viewing the resource, products, and job metrics. This allows to generate reports related to SDS processing statistics and product accountability on a defined interval.
The Helper Worker Services
Through the evolution of HySDS, there is already a set of special workers that handle the core processes. The set of workers serve out the non-PGE type jobs such as rule evaluation, e-mail notification, etc.. The key services include the following workers:
- Ingest Worker
- Triage Worker
The Ingest Worker automatically ingests files that are routinely and frequently delivered. It detects and retrieves ancillary data from the remote sites or the inbound staging area. PCM uses this function to catalog and store incoming data and extract the metadata. This includes the integrity verification of the ingested data files.
The Triage Worker is to detect early termination of compute resources and resubmit jobs. It also captures the artifacts of failed jobs after a set number of processing attempts.
PGE-PCM Interfaces
A set of PGE-PCM interfaces contains 20+ interfaces, one interface per PGE. PCM and PGE software complies to the interfaces based on each Interface and Control Flow Specification (ICS). Before PCM runs the PGE, PCM determines the location of required and alternate input files based on the selection rules. These input files include lower level product data, ancillary data, and engineering data files. PCM also determines and passes the required parameters to the PGE (e.g. pass and tile information, CRID, etc.). If all required conditions are met, PCM composes and generates the information into an interface file that is passed during the execution of PGE. This file contains parameters, input / output files, working directories, PGE software versions, etc.
When PCM executes a PGE, the following steps occur:
- PCM gathers the required information and generates the RunConfig file that PGE reads
- PCM localized inputs needed by PGE
- PCM initiates PGE with specified RunConfig
- PCM executes PGE and performs post-conditions defined
PCM tracks and monitors the status while PGE is processing. Once the PGE exits, PCM handles the exit conditions. There are two exit scenarios.
- Success
- Failure
If the PGE exits successfully, PCM handles the post-conditions including ingestion of the product granule, staging the product to outbound staging areas for CNES and PO.DAAC, update data accounting, and proceeding to the next processor in the pipeline (if not the end of the pipeline).
If the PGE fails, PCM updates the processing status, moves the processing artifacts to the triage area, and either retries the job or notifies to operators.
PCM Granule Boundaries Estimation (GBE) Database
PCM uses the GBE to do pass-based, tile-based, and cycle-based processing. Specifically, the GBE PGE reads the MOE file, the Orbit Revolution File, the Along Track Mapping, the Cross Track Mapping, and the Granule Overlap Amount. The GBE PGE uses the times in the Orbit Revolution File and the pass and tile boundaries in the Along Track Mapping and Cross Track Mapping to estimate when the instrument data will cover the geographical regions of the cycles, passes, and tiles. It also calculates sets of estimates including a percentage of overlap as given in the Granule Overlap Amount, directed by ADT. The output file contains the actual beginning and ending time estimates for the cycles, passes, and tiles in the along-track direction, and also the sets of overlap start and end times per each product. PCM uses these time-position mappings to:
- Look up the RangeBeginning and RangeEnding times to give to the L0B PGEs,
- Find all L0A_Prime files that overlap with the given RangeBeginning and RangeEnding times
- Associate the corresponding granule pass or tile with the given RangeBeginning and RangeEnding times for cataloging and file naming.
External Interface to CNES, GDS, and PO.DAAC
GDS and CNES push the incoming data files to SDS. PCM automatically monitors, detects, and ingests the defined data files at the inbound staging areas. The data files are checked for integrity before they’re ingested into PCM.
In support of the bulk reprocessing, the inputs (e.g., L0 products, ancil/aux data, etc) are retrieved from PO.DAAC. These inputs are deposited in the designated inbound staging area and are subject to the same ingest process.
For archival purpose, PCM makes available the generated data products at the rolling storage and life of instance storage location, notifying the location of the products to PO.DAAC. For data exchange purpose with CNES, PCM make available selected data products at the archive staging location. The CNES and PO.DAAC pull the files to their systems. PCM interfaces to PO.DAAC using Cumulus CNM (Cloud Notification Mechanism) send and recieve messages. The details of the interface mechanisms are defined below:
For products generated by CNES, PCM delivers these products to the DAAC using the following process:
- Products are placed in the archive staging location by CNES.
- PCM generates a CNM-S message which indicates the location of the product. This message is sent to DAAC using AWS Kinesis.
- DAAC ingests the product and generates a CNM-R message, indicating success or failure of ingestion. This message is sent to PCM using an SNS topic which is connected to a AWS Lambda.
- PCM processes and stores CNM-R messages. These messages are cataloged and later queried for activities such as bulk reprociessing, where a large number of previously ingested products must be gathered from the DAAC.
For products generated by the PCM, messages are delivered to both the DAAC and CNES using the following process:
- PCM generates products
- PCM creates and copies tar’d SDP datasets to the PCM Archive Staging bucket outgoing directory.
- PCM creates CNM-S message for product
- PCM immediately creates message and sends to PO.DAAC
- PO.DAAC ingests tar file
- PO.DAAC returns a CNM-R msg of successful/failed ingestion to PCM SDS via an AWS message service
- For success msgs, PCM updates catalog
- For failed msgs, PCM SDS troubleshoots
- Every X minutes, PCM creates a single CNM-S msg with all the tar files placed in the Archive Bucket outgoing directory and places the CNM-S msg in the PCM Msg Bucket CNM-S directory
- CNES polls the PCM Msg Bucket CNM-S directory every Y minutes and pulls the msg(s).
- CNES deletes the message
- CNES pulls the tar files identified in a msg to CNES
- CNES creates one or two CNM-R messages with the success and/or failure of the tar copying activity and places the msg into the PCM Msg Bucket CNM-R directory.
- CNES notifies distribution center of product(s)
- PCM immediately creates message and sends to PO.DAAC
Metrics & Data Accounting
PCM manages an ElasticSearch database for tracking input products, intermediate products, and all reprocessing campaigns. Metrics collected by the PCM system include:
- Quantity of products - How many products produced? How many received packets and gaps in packets?
- Continuity of products - How many files are missing or were not created?
- Latency of products - How much time to generate outputs?
Metrics are ingested from a variety of sources into the Metrics server's database. Each metrics source generates JSON file with relevant information
- Job runner on worker node instance
- Messages received from DAAC
- Filenames of products received from CNES
- PGE output
The JSON messages generated from these sources are simple, each with a filename key tying metrics from different sources to a single product. For example:
{
"filename": "",
"totalPackets": 0, // based on total_packets
"totalPacketsPriorToDiscard": 0, // based on total_packets - bad_pus - bad_ccsds
"invalidChecksumDiscardedPackets": 0, // based on bad_ccsds + bad_pus
"invalidCcsdsHeaderDiscardedPackets": 0, // based on bad_ccsds
"totalGaps":0, // based on total_packet_gaps
"gapList": [ { "gapNumber": 0, "lastFrameBeforeGap": "", "nextFrameAfterGap": "" }, // based on total_packet_gaps, packet_gap_time, time format is YYYY-MM-DDThh:mm:ss.sssZ
{ "gapNumber": 1, "lastFrameBeforeGap": "", "nextFrameAfterGap": "" },
{ "gapNumber": 2, "lastFrameBeforeGap": "", "nextFrameAfterGap": "" }, etc... ],
}
Information is sent to the Metrics server by these sources, stored in ElasticSearch, and made available in real-time to operators via Kibana. Metric reports are generated by querying the metrics ElasticSearch database and building an XML from the resulting data. Reports can be delivered by email or written to a location as needed. Reports generated at a configurable period such as daily, weekly, or monthly include:
- Data loss
- Results generated by PGE to pass via metadata to PCM
- PCM produces data loss summary report
- Data Accountability
- Data Product Latency
- SDS system statistics
- Product delivery to PO.DAAC & CNES
Customized Operator Interfaces
The inherited PCM core capability provides helpful and easy-to-view web interfaces including the following:
- Web application that provides browse, facet-search, and query of data and metadata.
- Web application that provides tracking of jobs and their status.
- Metrics Dashboard: web application via Kibana dashboard that provides metrics on processing and celery jobs, celery workers, and Verdi workers,
PCM leverages the inherited user interfaces and customizes as needed.
PCM Deployment
Multiple concurrently operational pipelines for specific processing scenarios including Forward Processing (Persistent), Bulk Processing (Pre-defined input), and ADT Processing (Temporary). Pipelines are deployed in separate AWS accounts. This allows for isolation of permissions, costing, AWS account limits. Certain resources, such as S3 buckets, are shared between accounts. Permissions for read/write access are specific to account and are configureable. PCM's deployment and development process is described in the following diagram:
This diagram shows the interactions developers have with with the PCM DEV cluster and how that deployed system flows to I&T and OPS deployments. In this diagram PCM is using Terraform as the PCM Cluster Launcher. PCM's process for any new, operational deployment, is as follows:
- System engineer notifies SA team of plan to deploy PCM. SA team provides needed cloud credentials to deploy PCM configuration.
- Deployment engineer configures deployment by running PCM deployment configuration tool. Tool generates PCM deployment config.
- Can optionally re-use a pre-generated configuration.
- Terraform runs, deploying AWS infrastructure and provisioning PCM components within minutes. Pre-build AMIs are used for EC2 instantiation.
- PCM pulls docker images for PGEs
- Terraform completes, providing operator with URLs to operator web interfaces to verify deployment.
When deployed, PCM supports using Amazon EC2 Spot Instance workers:
- System Engineer specifies price policy (willing to pay per hour).
- If Spot market price exceeds the specified price, spot instances are terminated with a 2-minute warning.
- Spot termination likelihood is directly tied to price.
Design for Spot termination notice:
- spot_termination_detector is notified of termination by AWS
- Sends graceful shutdown signal to node’s docker container
- Submits event log that node was marked for termination
- Incomplete jobs are tracked in the resource manager as never returning due to timeout tracked in RabbitMQ
Incomplete jobs are re-run on a different EC2 node
Glossary
Acronym/Term | Description |
|---|---|
| ARIA | Advanced Rapid Imaging and Analysis |
| ARIA-SG | Advanced Rapid Imaging and Analysis (Earth Observatory of Singapore) |
| AWS | Amazon Web Services |
| DAAC | Data Active Archive Center |
| GCP | Google Cloud Platform |
| GDS | Ground Data System |
| HDDR | High Density Digital Recorder |
| HTTP/HTTPS | HyperText Transfer Protocol / HyperText Transfer Protocol Secure |
HySDS | Hybrid Cloud Science Data System |
| IDP | Interim Digital SAR Processor |
| JPL | Jet Propulsion Laboratory |
| JSON | JavaScript Object Notation |
NASA | National Aeronautics and Space Administration |
| NISAR | NASA-ISRO Synthetic Aperture Radar Mission |
PGE | Product Generation Executor/Executable |
| SAR | Synthetic Aperture Radar |
SDS | Science Data System |
| SMAP | Soil Moisture Active Passive Mission |
| SSL | Secure Sockets Layer |
| SWOT | Surface Water Ocean Topography Mission |
| WVCC | Water Vapor Cloud Climatology |
| XML | eXtensible Markup Language |
References
Cohen and P. Agram, "Leveraging the usage of GPUs in SAR processing for the NISAR mission," 2017 IEEE Radar Conference (RadarConf), Seattle, WA, 2017, pp. 0492-0496. 10.1109/RADAR.2017.7944253
LaHaye, Nick. (2012). Pleiades and OCO-2: using supercomputing resources to process OCO-2 science data final report. NASA Undergraduate Student Research Program (USRP), Pasadena, California.
Wu, C & Barkan, B & Huneycutt, B & Leang, C & Pang, S. (1981). An introduction to the interim digital SAR processor and the characteristics of the associated Seasat SAR imagery.
Document Change Log
| Version | Date | Comment |
|---|---|---|
| Current Version (v. 6) | May 29, 2019 18:16 | Stirling Algermissen |
| v. 5 | May 28, 2019 18:36 | Stirling Algermissen |
| v. 4 | May 28, 2019 18:27 | Stirling Algermissen |
| v. 3 | May 28, 2019 18:12 | Stirling Algermissen |
| v. 2 | May 28, 2019 16:30 | Stirling Algermissen |
| v. 1 | May 24, 2019 18:10 | Stirling Algermissen |
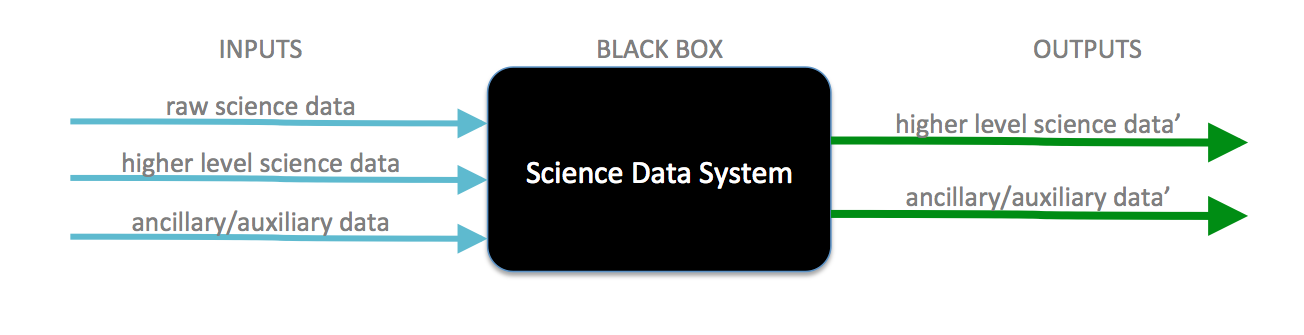
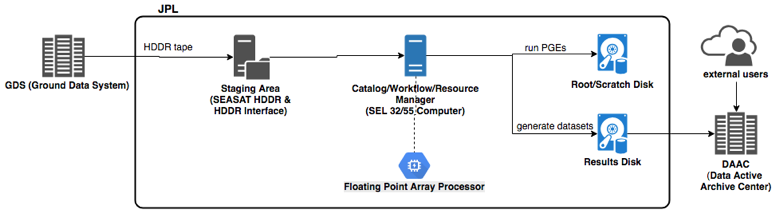
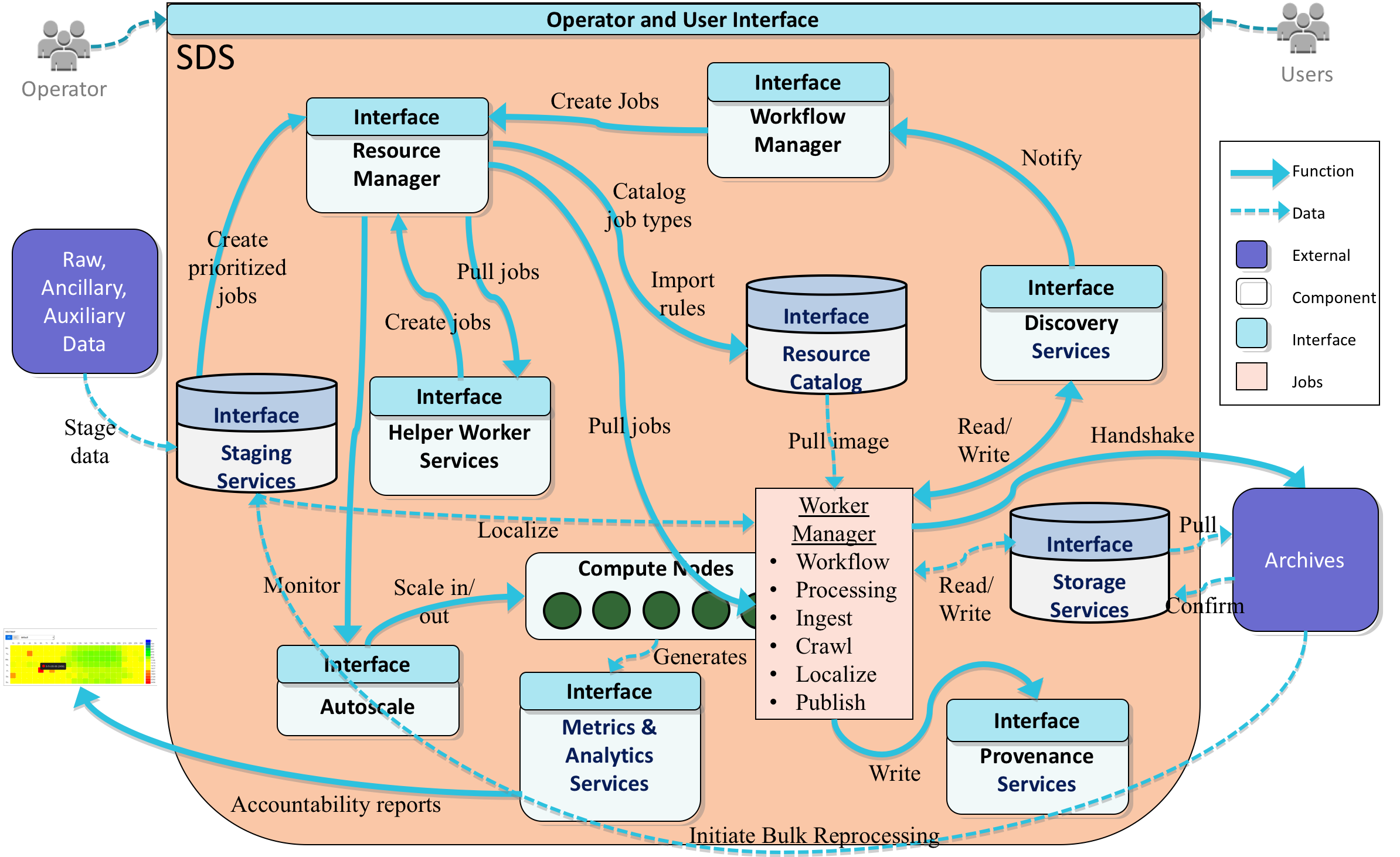
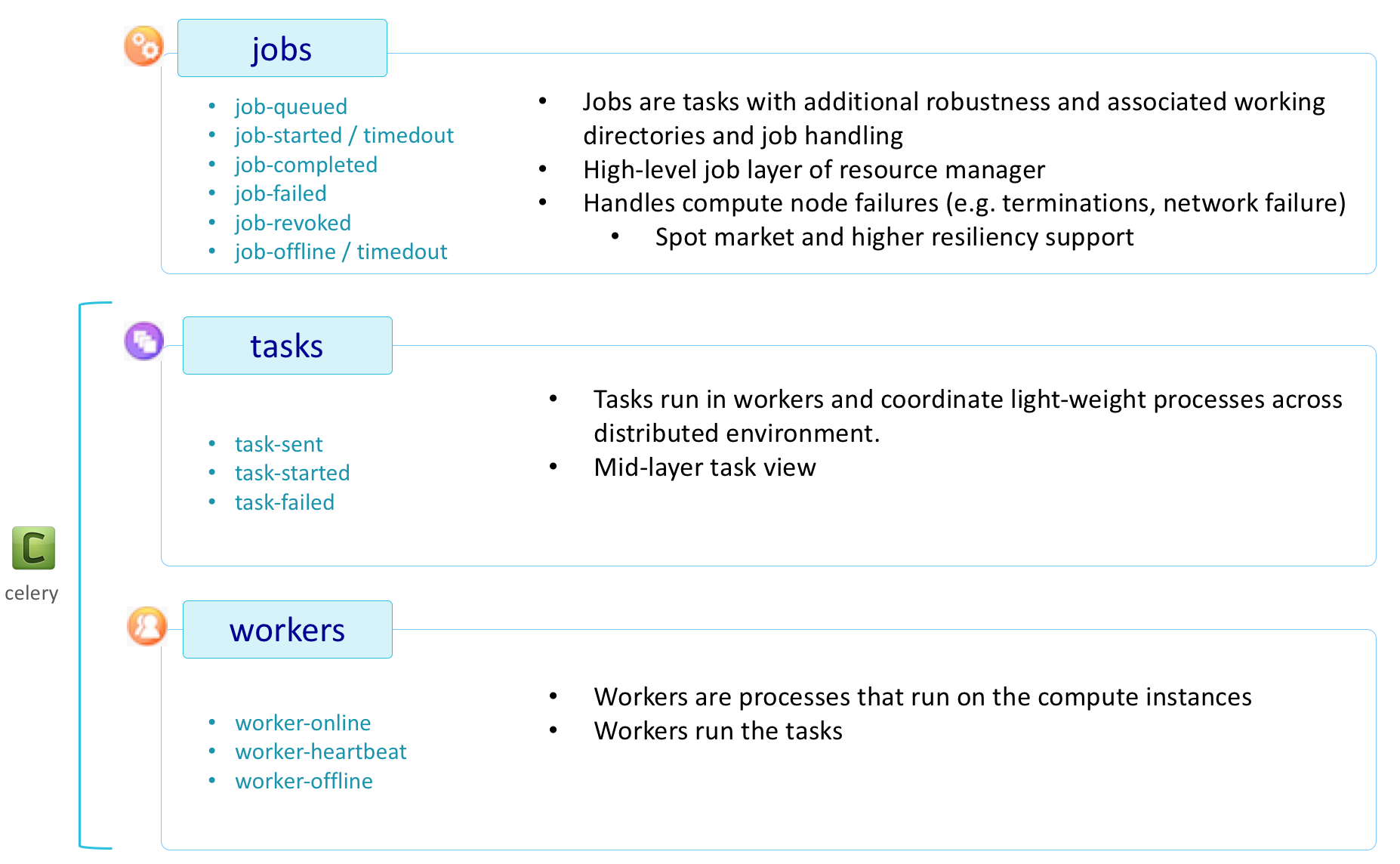
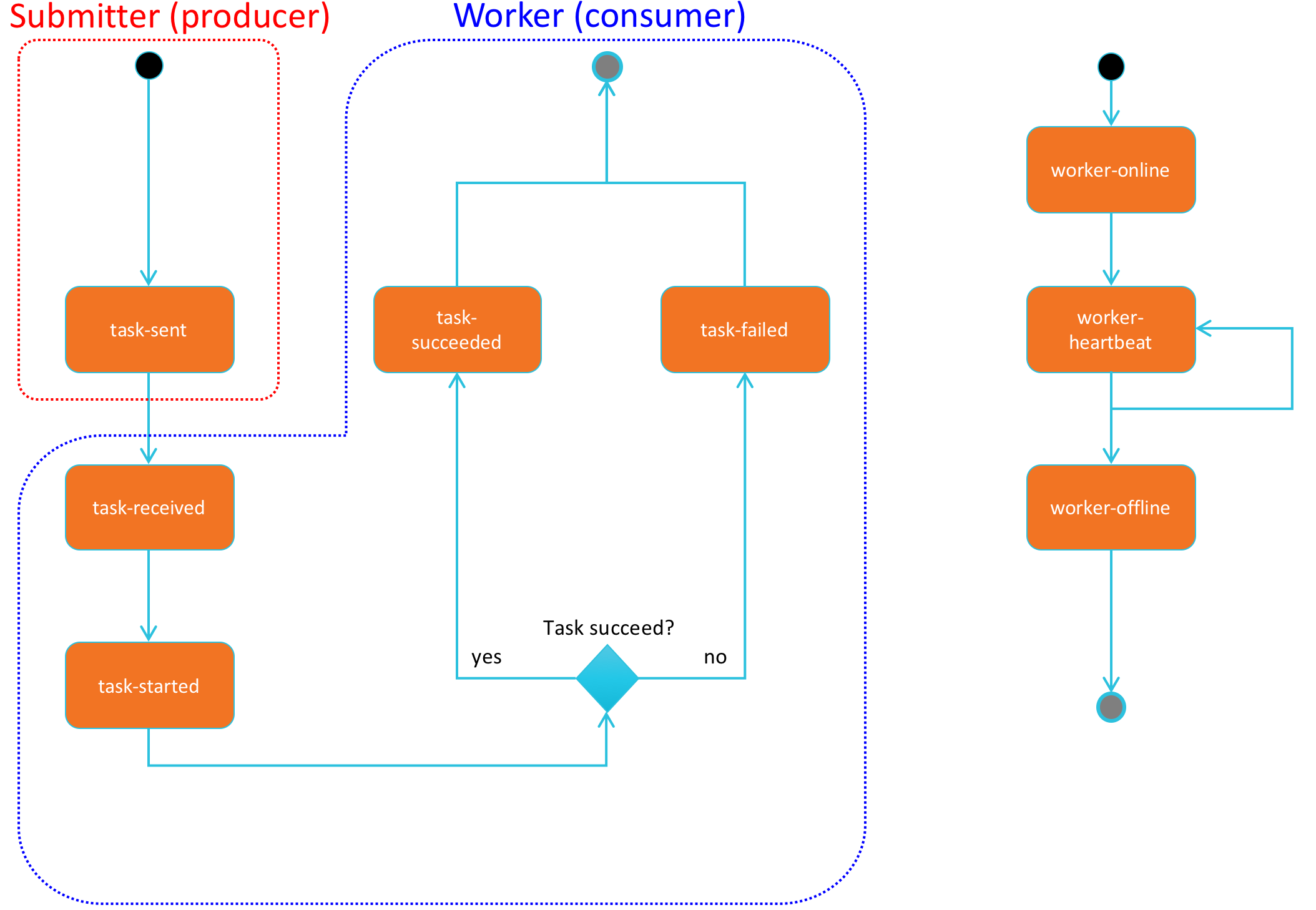
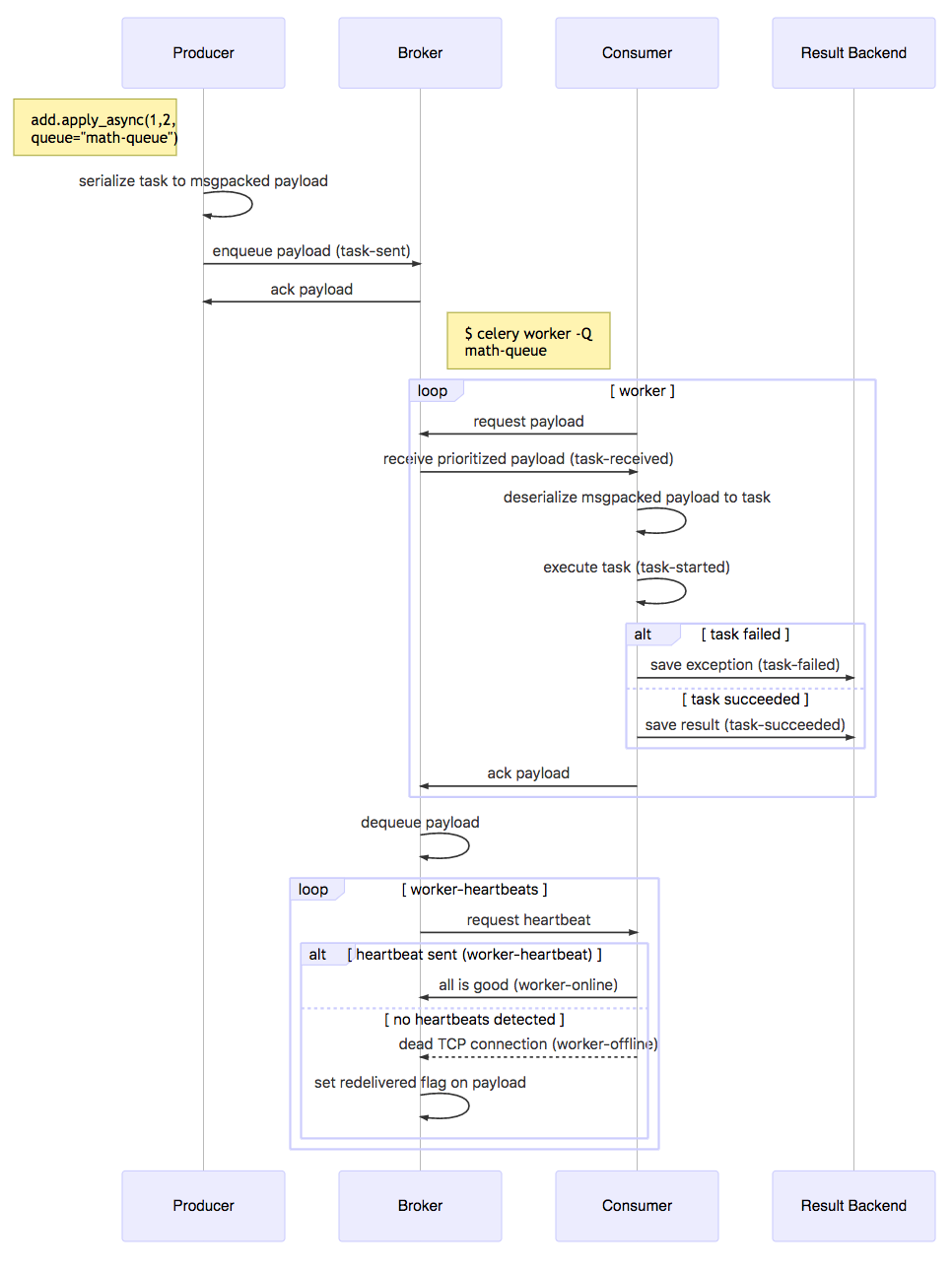
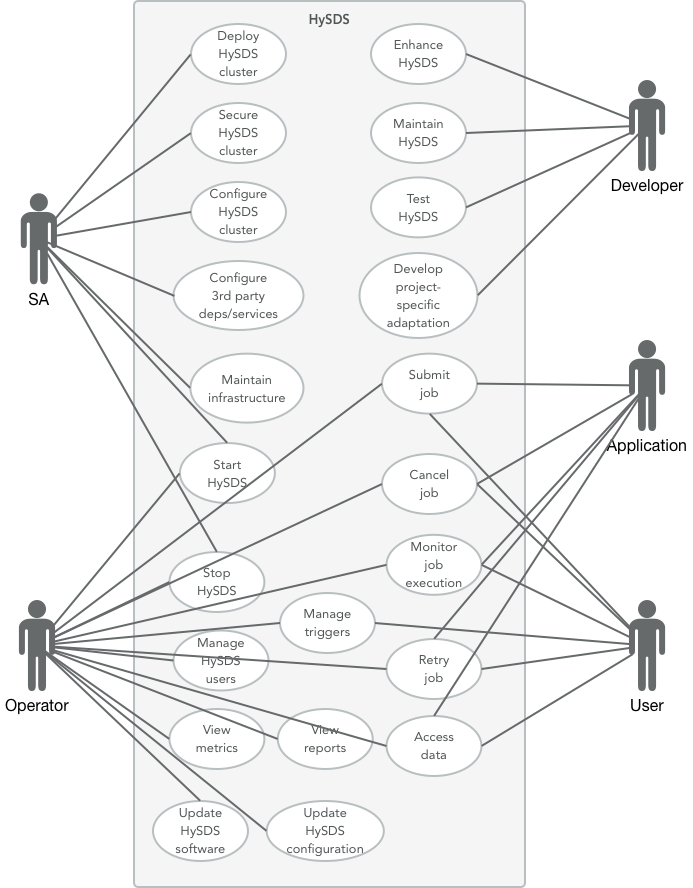
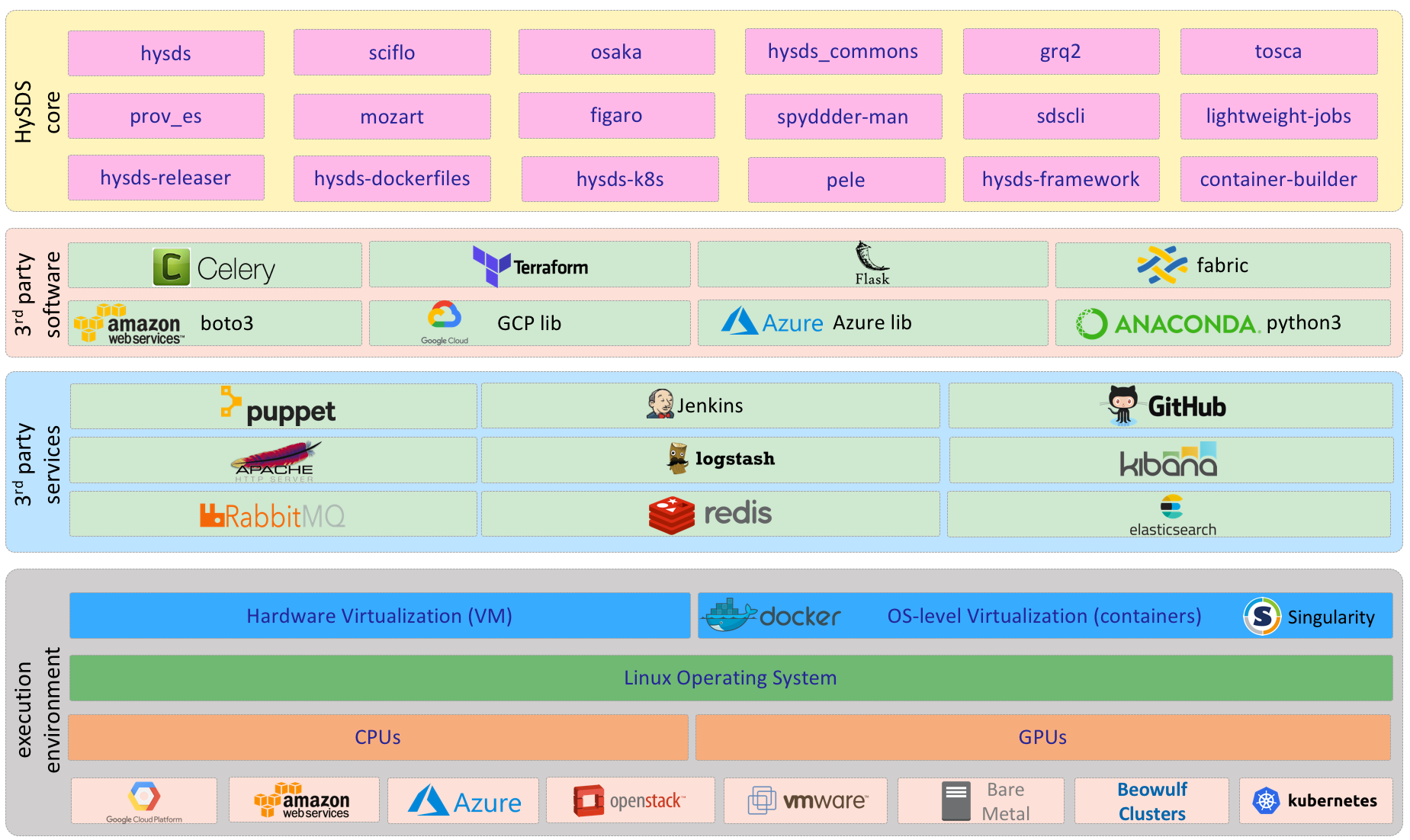
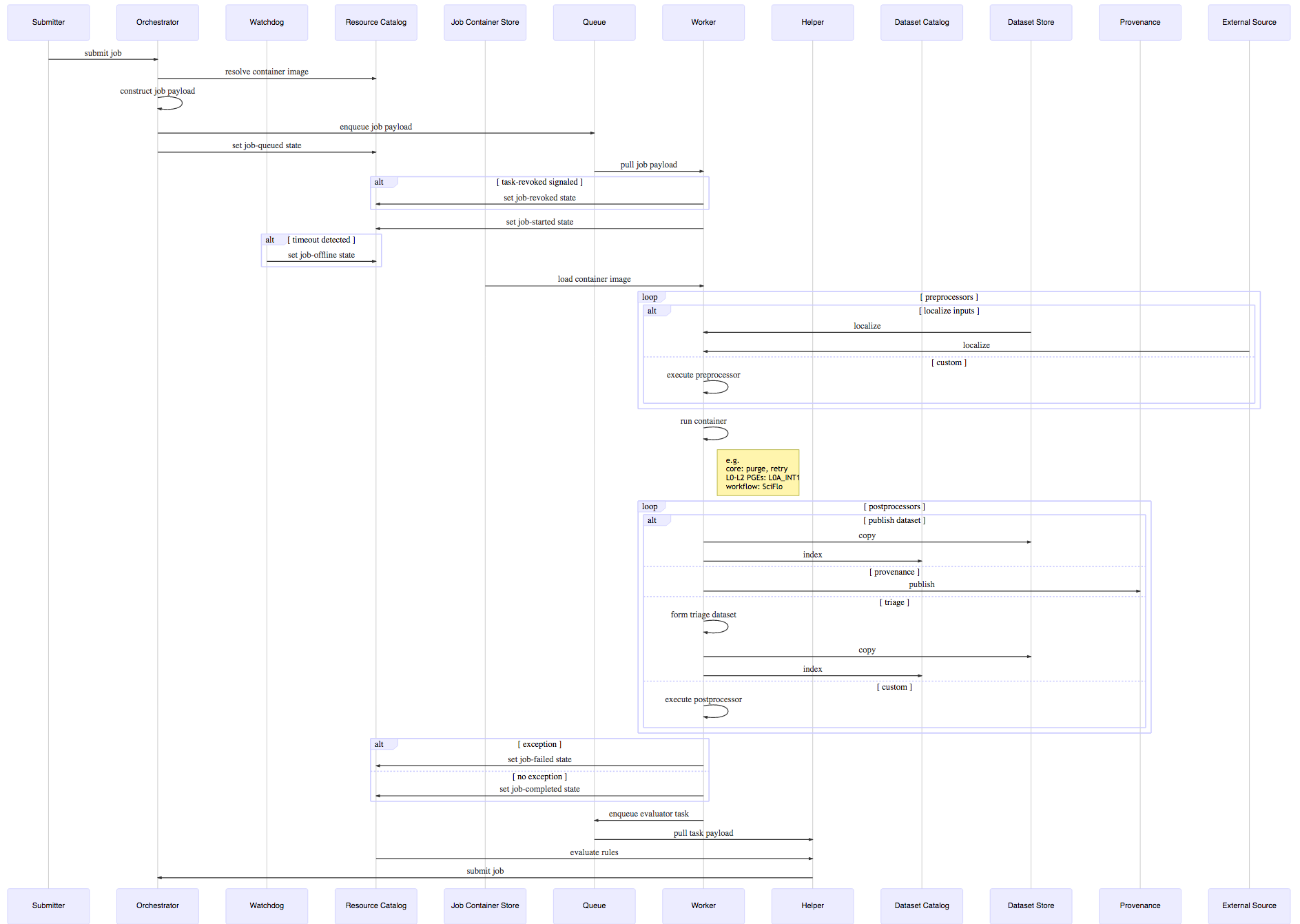
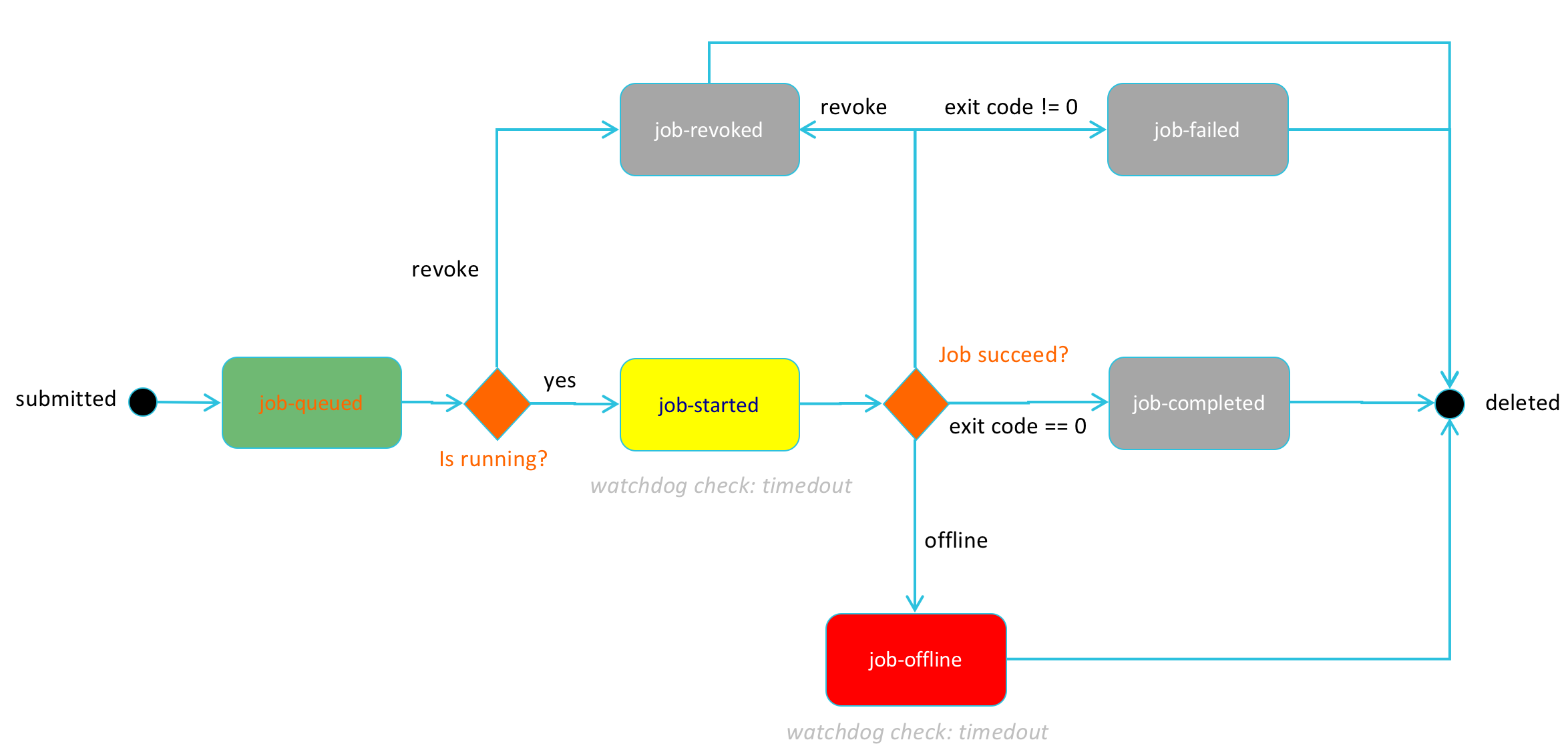
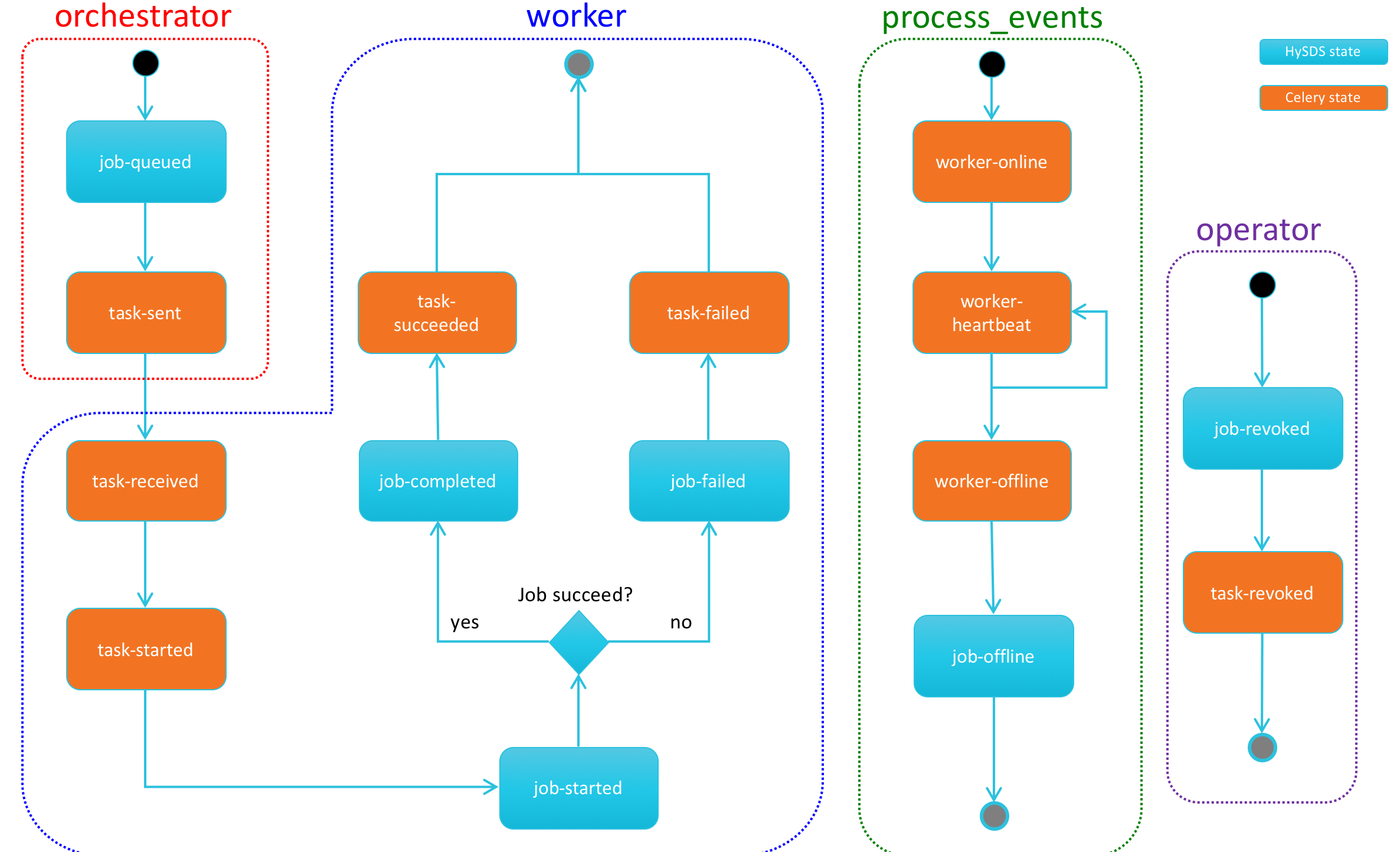
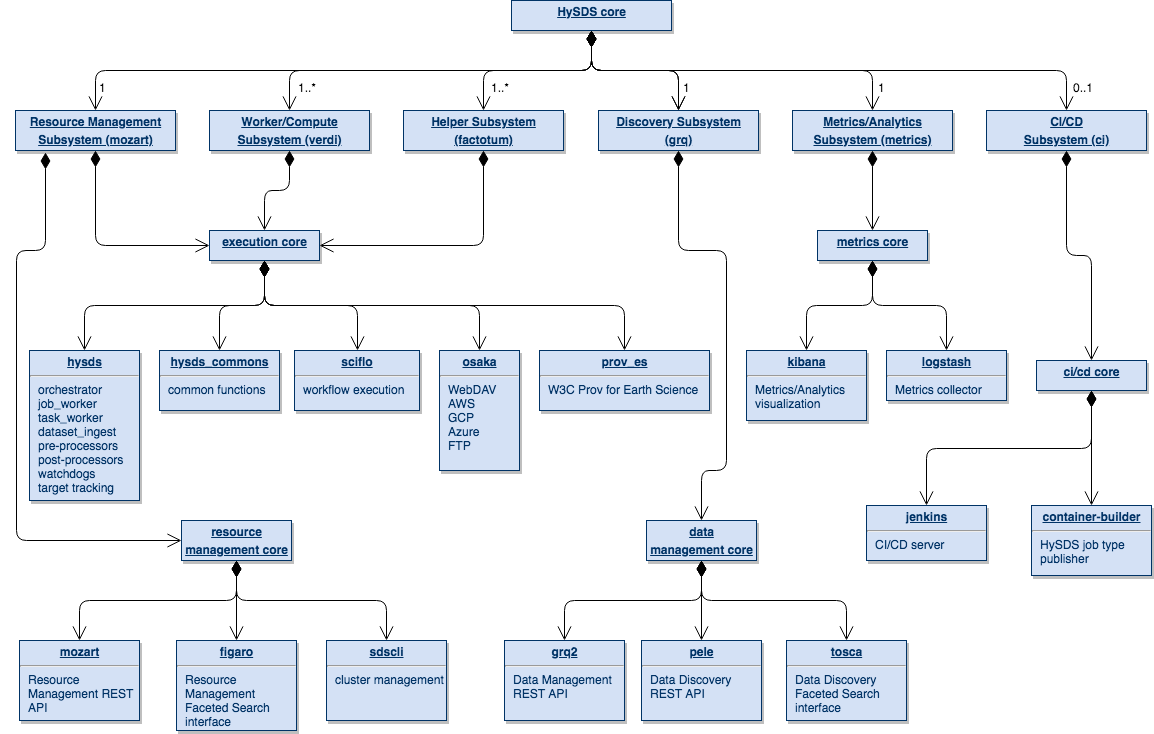
.png?version=1&modificationDate=1558721112916&cacheVersion=1&api=v2)